Antimicrobial resistance (AMR) is a global public health crisis that requires coordinated efforts between countries and international organizations. Public health institutions, including the CDC and WHO, recognize AMR as one of the most important threats to public health of our time. Rates of multidrug-resistant (MDR) bacterial isolates, including extended-spectrum βlactamases (ESBLs), carbapenem-resistant Enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa, are on the rise worldwide.1–4
The most common mechanism for development of multidrug-resistance in Gram-negative bacteria is via horizontal transfer of plasmids carrying resistance genes, particularly carbapenemases.5 Carbapenemases are members of the molecular Class A, B, and D β-lactamases and can be organized further into enzyme families.6 Class A and D β‐lactamases work by a serine-based hydrolytic mechanism. Klebsiella pneumoniae carbapenemases (KPCs) are the most prevalent of the class A group, whereas oxacillin-hydrolyzing–type (OXA) carbapenemases are the most prevalent class D carbapenemases.7 Class B enzymes are metallo-β-lactamases and include the VIM (verona integron-mediated metallo-β-lactamase), imipenem metallo-β-lactamase (IMP) (active on IMP), and New Delhi metallo-β-lactamase (NDM) families (among others). Carbapenemase gene detection is a critical component of surveillance at all levels—local, national, and international.
Much of the burden of AMR in resource limited settings remains unknown. In Nicaragua, no prior published studies have characterized AMR among clinical isolates. From 2014 to 2017, 334 Gram-negative rod (GNR) isolates were sent to Centro Nacional de Diagnóstico y Referencia (CNDR) for confirmation of carbapenemases; 268 were processed and 66 were nonviable (Supplemental Table 1). Isolate distribution by year is shown in Supplemental Table 1. Of the 268 isolates tested, 231 isolates were defined as being resistant to imipenem by disk diffusion. This pilot study examined 100 selected MDR-GNR isolates from throughout the country to determine the genetic relatedness of resistant strains and better characterize the mechanisms of resistance.
Isolates from sterile sites collected from 2014 to 2017 from hospitals throughout Nicaragua were originally sent to the CNDR in Managua. Isolates were obtained from patients ranging in age from 1 day old to 90 years old and from a variety of hospital departments, including the neonatal intensive care unit (ICU), pediatric and neonatal floors, and adult ICU. The majority of the isolates were cultured from blood (81%) with the remainder cultured from pleural fluid (7%), cerebrospinal fluid (CSF) (4%), peritoneal fluid (2%), bronchial lavage (2%), and source unknown (4%) (Supplemental Table 2). Isolates were identified at the CNDR using the Vitek 2 (bioMérieux, Marcy l’Etoile, France). Isolates with carbapenem resistance identified by E-test, with or without confirmed resistance to additional antibiotic classes, were stored in media supplemented with 25% glycerol. Hundred suspected multidrug-resistant Gram-negative rods (MDR-GNRs) representing the most common species (Escherichia coli, K. pneumoniae, A. baumannii, and P. aeruginosa) were shipped to Emory University for further characterization. Of the 100 isolates, 16% were P. aeruginosa, 30% were A. baumannii, and 54% were Enterobacteriaceae. The majority of Enterobacteriaceae were K. pneumoniae (50/54, 93%) and remainder were E. coli (4/54, 7%).
Susceptibility testing of all isolates to appropriate carbapenems for each species was performed by disk diffusion according to Clinical and Laboratory Standards Institute.8 Resistance to carbapenems was confirmed for 99 of the 100 isolates. One P. aeruginosa isolate was found to be susceptible to all carbapenems tested. All isolates were found to be susceptible to colistin by E-test (Supplemental Table 2). Although colistin testing by gradient diffusion has limitations,8 many clinical microbiology laboratories use this methodology. In addition, our isolates had very low minimum inhibitory concentrations (none bordering a resistant phenotype or colonies inside the zone of inhibition) and it is, therefore, unlikely that any resistant isolates were missed by this method (Supplemental Table 2).
Enterobacteriaceae isolates were tested for the presence of ESBLs using a combination disk diffusion method (ceftazidime + ceftazidime-clavulanate and cefotaxime + cefotaxime-clavulanate).9 Presence of an ESBL was found in 21/50 (42%) of the K. pneumoniae isolates, but none of the E. coli isolates. All isolates (Enterobacteriaceae, A. baumannii, and P. aeruginosa) were tested for carbapenemase production by the modified carbapenem inactivation method (mCIM).9 All 54 Enterobacteriaceae, 11/16 (68%) P. aeruginosa, and 1/30 (3%) of A. baumannii were found to produce a carbapenemase by the mCIM. Results are shown in Table 1 and Supplemental Table 2.
Summary of findings for all isolates
| Carbapenem resistance* | Extended-spectrum β-lactamases | Carbapenemase | |||
|---|---|---|---|---|---|
| Species | Ertapenem | Doripenem | Imipenem | Producer† | Producer (%)‡ |
| Klebsiella pneumonia, n = 50 | 49 (98%)§ | 48 (96%)§ | 49 (98%)§ | 21 (42%) | 50 (100%) |
| Escherichia coli, n = 4 | 4 (100%) | 4 (100%) | 4 (100%) | 0 (0%) | 4 (100%) |
| Pseudomonas aeruginosa, n = 16 | ND | 15 (94%)‖ | 14 (87%)‖ | ND | 11 (68.8%) |
| Acinetobacter baumannii, n = 30 | ND | 30 (100%) | 30 (100%) | ND | 1 (3%)¶ |
ND = not tested.
* Determined by disk diffusion.
† Determined using a combination disk diffusion method (ceftazidime + ceftazidime–clavulanate and cefotaxime + cefotaxime–clavulanate).
‡ Determined by the modified carbapenem inactivation method (mCIM) phenoptypic test for the production of carbapenemase.
§ Remaining isolates were intermediate to ertapenem, doripenem, and meropenem.
‖ One P. aeruginosa was susceptible to all carbapenems tested and one was intermediate to imipenem.
¶ Note: mCIM has been found to be less accurate for A. baumannii.
Bacterial DNA was extracted using an eMag® instrument (bioMérieux), and all isolates were screened for the presence of β-lactamase resistance genes blaKPC, blaOXA-48, blaVIM, blaIMP, and blaNDM as previously described.10,11 Table 2 details the distribution of carbapenemase genes by each bacterial species. All Enterobacteriaceae isolates carried a carbapenemase gene with the vast majority 50/54 (93%) being positive for the blaNDM; 46/50 (92%) K. pneumoniae and 4/4 (100%) E. coli. The blaKPC was found in 4/50 (8%) of the K. pneumoniae isolates. Acinetobacter baumannii and P. aeruginosa isolates were less likely to have a carbapenemase gene identified compared with the Enterobacteriaceae; 11/16 (68.8%) of the P. aeruginosa and 10/30 (33%) of A. baumannii isolates carried a carbapenemase gene. Pseudomonas aeruginosa isolates carried blaVIM and blaIMP carbapenemase genes only, with the majority of these isolates (10/11, 91%) having both. The 10/30 (33%) of A. baumannii isolates with a carbapenemase gene were identified as blaNDM. PCR confirmed the results of the mCIM for all Enterobacteriaceae and P. aeruginosa isolates. The mCIM results were less reliable for use with A. baumannii isolates as seen by others.12
Distribution of carbapenemase genes by bacterial species
| Species | blaKPC (%) | blaNDM (%) | blaVIM (%) | blaIMP (%) | blaOXA-48 (%) |
|---|---|---|---|---|---|
| Klebsiella pneumoniae, n = 50 | 4 (8) | 46 (92) | 0 (0) | 0 (0) | 0 (0) |
| Escherichia coli, n = 4 | 0 (0) | 4 (100) | 0 (0) | 0 (0) | 0 (0) |
| Pseudomonas aeruginosa, n = 16 | 0 (0) | 0 (0) | 11 (68.8) | 10 (62.5) | 0 (0) |
| Acinetobacter baumannii, n = 30 | 0 (0) | 10 (33) | 0 (0) | 0 (0) | 0 (0) |
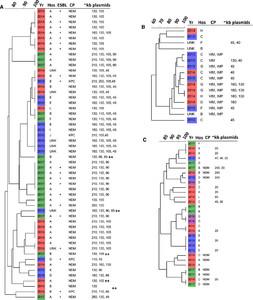
Genetic relatedness and plasmid sizes were determined by PFGE using the CHEF-Mapper (Bio-Rad, Hercules, CA) and analyzed with BioNumerics v. 7.6 (Applied Maths, Saint-Martens-Latem, Belgium).13–16 PFGE analysis revealed patterns of clustering among isolates. Isolates appeared to cluster by three factors: plasmid profiles, year of culture, and health care setting (Figure 1). Clustering was particularly apparent for K. pneumoniae isolates (Figure 1A); however, more plasmids were identified within these strains.



Cluster analysis of PFGE patterns (data not shown), carbapenemases, and plasmids of Enterobacteriaceae (A), Pseudomonas aeruginosa (B), and Acinetobacter baumannii (C) isolates from 2014 to 2017 (Yr) shown in red, blue, purple, and green, respectively. Percent similarity of PFGE bands at top of dendrogram. Size of plasmids is approximate kb.
Citation: The American Journal of Tropical Medicine and Hygiene 102, 2; 10.4269/ajtmh.19-0561
This study reveals very high rates of NDM-carbapenemase genes in MDR-GNRs from hospitals throughout Nicaragua. Prior studies have shown the presence of NDM-1 in Latin and Central America, but there is limited evidence regarding the prevalence of this resistance mechanism in most of Central America, and particularly in Nicaragua.17–19 The burden of MDR-GNR may be underestimated as approximately 38% of the isolates were either nonviable or not received by the CNDR. PFGE analysis revealed clusters of isolates by year of isolation, health care setting, and approximate plasmid size, suggesting genetic relatedness. Available clinical data for the 100 tested isolates were limited. However, only strains isolated from sterile sites were selected for this study, indicating the clinical relevance of these data.
Further research is needed to examine clinical and epidemiologic factors associated with MDR isolates and to determine patterns of acquisition, as well as high-risk exposures or comorbidities associated with MDR infection in Nicaragua. In settings with limited health care resources, cost-effective management and surveillance strategies are warranted to control transmission and provide appropriate empiric therapy to individuals with bacterial infections to improve clinical outcomes. This study allows us to draw conclusions about the prevalence of certain types of carbapenemase among MDR-GNR isolates; however, further research is needed to determine the overall prevalence of MDR-GNR. A prospective study that collects epidemiologic and geographic information for isolates could help identify specific regions and populations within Nicaragua with higher rates of community MDR carriage. Determining the burden of highly resistant bacteria in Nicaragua is a crucial first step to guide interventions to limit further spread both locally and globally.
Acknowledgment:
We thank Janet Thonkulpitak for her help with the PFGE.
REFERENCES
- 1.↑
Centers for Disease Control and Prevention (CDC), 2013. Vital signs: carbapenem-resistant Enterobacteriaceae. MMWR Morb Mortal Wkly Rep 62: 165–170.
- 2.
Gupta N, Limbago BM, Patel JB, Kallen AJ, 2011. Carbapenem-resistant Enterobacteriaceae: epidemiology and prevention. Clin Infect Dis 53: 60–67.
- 3.
Evans BA, Hamouda A, Amyes SG, 2013. The rise of carbapenem-resistant Acinetobacter baumannii. Curr Pharm Des 19: 223–238.
- 4.↑
Grass J et al. 2018. Epidemiology of carbapenem-resistant Pseudomonas aeruginosa identified through the emerging infections program (EIP), United States, 2016–2017. Open Forum Infect Dis 5 (Suppl 1): S349–S350.
- 5.↑
Hardiman CA, Weingarten RA, Conlan S, Khil P, Dekker JP, Mathers AJ, Sheppard AE, Segre JA, Frank KM, 2016. Horizontal transfer of carbapenemase-encoding plasmids and comparison with hospital epidemiology data. Antimicrob Agents Chemother 60: 4910–4919.
- 6.↑
Bonomo RA, Burd EM, Conly J, Limbago BM, Poirel L, Segre JA, Westblade LF, 2018. Carbapenemase-producing organisms: a global scourge. Clin Infect Dis 66: 1290–1297.
- 7.↑
Queenan AM & Bush K 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20: 440–458.
- 8.↑
Matuschek E, Åhman J, Webster C, Kahlmeter G, 2018. Antimicrobial susceptibility testing of colistin – evaluation of seven commercial MIC products against standard broth microdilution for Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa and Acinetobacter spp. Clin Microbiol Infect 8: 865–870.
- 9.↑
Clinical and Laboratory Standards Institute (CLSI) 2017. Performance Standards for Antimicrobial Susceptibility Testing, 27th edition. Wayne, PA: M-100, CLSI.
- 10.↑
Rasheed JK, Kitchel B, Zhu W, Anderson KF, Clark NC, Ferraro MJ, Savard P, Humphries RM, Kallen AJ, Limbago BM, 2013. New Delhi metallo-β-lactamase-producing Enterobacteriaceae, United States. Emerg Infect Dis 19: 870–878.
- 11.↑
Kitchel B et al. 2013. Detection and evaluation of OXA-48 like carbapenemases by real-time PCR, Abstracts of the 53rd Meeting ICAAC, September 10–13, 2013, Denver, Colorado. Abstract D-1139.
- 12.↑
Simner PJ et al. 2018. Multicenter evaluation of the modified carbapenem inactivation method and the carba NP for detection of carbapenemase-producing Pseudomonas aeruginosa and Acinetobacter baumannii. J Clin Microbiol 56: e01369–17.
- 13.↑
Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ, 2006. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis 3: 59–67.
- 14.
Barton BM, Harding GP, Zuccarelli AJ, 1995. A general method for detecting and sizing large plasmids. Anal Biochem 226: 235–240.
- 15.
Selim S, El Kholy I, Hagagy N, El Alfay S, Aziz MA, 2015. Rapid identification of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. Biotechnol Equip 29: 152–156.
- 16.↑
Seifert H, Dolzani L, Bressan R, van der Reijden T, van Strijen B, Stefanik D, Heersma H, Dijkshoorn L, 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol 43: 4328–4335.
- 17.↑
López-Leal G et al. 2019. Unexplored genetic diversity of multidrug- and extremely drug-resistant Acinetobacter baumannii isolates from tertiary hospitals in Honduras. Microb Drug Resist 5: 690–695.
- 18.
Marquez-Ortiz RA et al. 2017. Genomic epidemiology of NDM-1-encoding plasmids in Latin American clinical isolates reveals insights intothe evolution of multidrug resistance. Genome Biol Evol 6: 1725–1741.
- 19.↑
Torres-González P, Bobadilla-Del Valle M, Tovar-Calderón E, Leal-Vega F, Hernández-Cruz A, Martínez-Gamboa A, Niembro-Ortega MD, Sifuentes-Osornio J, Ponce-de-León A, 2015. Outbreak caused by Enterobacteriaceae harboring NDM-1 metallo-β-lactamase carried in an IncFII plasmid in a tertiary care hospital in Mexico City. Antimicrob Agents Chemother 11: 7080–7083.