Introduction
Human immunodeficiency virus type 1 (HIV-1) is classified into three major groups (M, N, and O), in which HIV-1 group M is responsible for global pandemic with nine established HIV-1 subtypes and various circulating recombinant forms (CRFs) (www.hiv.lanl.gov). The Joint United Nations Program on HIV/AIDS (UNAIDS) estimated around 35.3 million people living with HIV-1 globally with 1.6 million deaths caused by acquired immunodeficiency syndrome (AIDS) and 2.3 million of new infections in by the end of 2012. In Southeast Asia, an estimated 3.9 million people are living with HIV-1, with 270,000 new HIV-1 infections and 220,000 AIDS-related deaths reported.1
The HIV-1 subtype B and CRF01_AE are the predominant genotypes in Southeast Asia in the past decades.2 More recently, HIV-1 CRF33_01B has been steadily expanding in the Asia Pacific region (Malaysia, Singapore, Indonesia, Australia, and Hong Kong3–7) and has caused infections among various risk groups, including the low-risk populations.8 Co-circulation of subtype B, CRF01_AE, and CRF33_01B has also led to the emergence of various recombinant variants.8 Recombinant lineages with identical recombination structure that caused epidemic spread are classified as CRFs,9 whereas individual viral isolates with a distinct recombination pattern are termed as the unique recombinant form (URF). To date, at least nine novel CRFs have emerged as the result of a recombination between various HIV-1 lineages in Southeast Asia: five in Malaysia,6,10–13 three in Thailand,14–16 and one in Singapore.17 Because of the extensive genetic diversity and broad distribution of CRFs among the HIV-1-infected population in Southeast Asia, the availability of a rapid and multi-target subtyping tool will be of importance for high-throughput detection of classical and novel HIV-1 genotypes circulating in the region.
Continual advances in nucleic acid testing have enabled molecular epidemiological surveillance of rapidly evolving human pathogens such as HIV-1 to be carried out. Real-time polymerase chain reaction (PCR) by TaqMan assay uses fluorescent probes that enables simultaneous identification of targeted gene(s) during PCR amplification. Real-time PCR technology is an appealing alternative to conventional PCR as a result of the combination of excellent sensitivity and specificity, low contamination risk, and ease of performance and speed.18 Availability of a multiplex real-time PCR tool provides rapid and accurate identification of HIV-1 genotypes in areas where the HIV-1 prevalence is high, allowing large-scale molecular surveillance to be conducted in the most resource effective manner.19–23 A subtyping strategy that takes into account the spatial diversity of the virus, if applied to prospective cohorts, can be used to track the movement or diffusion of the common and also atypical HIV-1 lineages over the large geographical vicinity. A multi-region subtyping assay can be used as a timely and powerful “CRF discovery” tool in a region where novel CRFs have been continuously emerging.22 In this study, we developed an accurate multiplex real-time PCR assay for rapid detection of classical and recently emerging HIV-1 genotypes in Southeast Asia, and discussed its application in studying clinical and field samples in a setting of high HIV-1 genetic complexity. The method described in this study highlights the importance of active molecular surveillance necessary to understand the spatial and temporal dynamics of HIV-1,8 which may in turn inform the public health experts of effective intervention strategies, including vaccine selection and development.
Materials and Methods
Sample collection and nucleic acid extraction.
A total of 180 clinical plasma specimens from HIV-1-infected individuals from the University Malaya Medical Center (UMMC), Kuala Lumpur collected between 2008 and 2012 were screened for HIV-1 genotypes. The study was approved by the UMMC Medical Ethics Committee. Standard and multilingual consent forms were used and written consent was obtained from all study participants. The HIV-1 viral RNA was purified from plasma samples using the NucliSENS EasyMAG automated platform (bioMerieux, Durham, NC) following the manufacturer's protocol.
Primers and probes design.
Complete or near full-length genomes of CRF01_AE (N = 596) and subtype B (N = 189) of Asian origin were retrieved from the Los Alamos National Laboratory (LANL) HIV sequence database (www.hiv.lanl.gov). The sequences were aligned using the web-based multiple sequence alignment program MAFFT24 and manually edited where necessary. Consensus sequences were built and compared for each subtype. Primers and probes for real-time PCR were designed using Primer Express Software v2.0 (Applied Biosystems, Foster City, CA) at three different targeted regions (gag, pol, and env regions) based on the genome organization of HIV-1 subtype B, CRF01_AE, and the mosaic recombinant descendants of CRF33_01B, CRF53_01B, CRF54_01B, and CRF58_01B (Figure 1). The three genetic regions targeted in this assay were as follows: 1) the gag p24 region for CRF01_AE and CRF01_AE-origin segments in CRF33_01B, CRF53_01B, and CRF58_01B; 2) the pol protease-reverse transcriptase (pro-RT) region for subtype B and subtype B-origin segments in CRF33_01B, CRF54_01B, and CRF58_01B; and 3) the env gp41 region for subtype B and subtype B-origin segment in CRF58_01B. Newly designed primers and probes are listed in Table 1.



Genome organizations of full-length human immunodeficiency virus type 1 (HIV)-1 subtype B, CRF01_AE, CRF33_01B, CRF53_01B, CRF54_01B, and CRF58_01B. The HIV-1 subtype B and CRF01_AE, shown respectively in dark and light shades, are the putative parental genotypes for the various recombinant lineages (CRF33_01B, CRF53_01B, CRF54_01B, and CRF58_01B) reported previously. Multiplex real-time polymerase chain reaction (PCR) primers and TaqMan probes designed at three different genetic regions (p24, pro-RT, and gp41) are indicated by dashed vertical lines. Real-time PCR results or yield for each genetic region, as indicated by positive or negative amplification, will be used to determine the HIV-1 genotypes.
Citation: The American Society of Tropical Medicine and Hygiene 92, 3; 10.4269/ajtmh.14-0681
Primers and probes designed for the real-time PCR HIV-1 genotyping assay
| Sequence 5′ to 3′* | HXB2 location (nt) | |
|---|---|---|
| p24 region | ||
| Outer Primers | ||
| Forward | GGTGCGAGAGCGTC | 793–806 |
| Reverse | ATGCTRTCATCATYTCTTC | 1819–1837 |
| Inner Primers | ||
| Forward | ATGGGTRAARGTARTAGAAGAAAAGGG | 1251–1277 |
| Reverse | CTGCCTGRTGYCCYCCCACTA | 1358–1378 |
| Probe | FAM-CCCACAAGATYTAAA-MGB | 1329–1343 |
| pro-RT region | ||
| Outer Primers | ||
| Forward | CAGGAGCWGATGAYACAGT | 2329–2347 |
| Reverse | AATAYTGGRGTATTRTATGGA | 2711–2731 |
| Inner Primers | ||
| Forward | YCAGMTTGGNTGYACTTTAAATTTYCCC | 2525–2552 |
| Reverse | TTTYCCTTCYTTYTCCATTTCKG | 2665–2687 |
| Probe | VIC-ACARTGGCCATTRACAGA-MGB | 2615–2632 |
| gp41 region | ||
| Outer Primers | ||
| Forward | TGTTGCAACTCACAGTCT | 7918–7935 |
| Reverse | TGARTATCCCTKCCTAAC | 8346–8363 |
| Inner Primers | ||
| Forward | TGGGGNTGYTCTGGAAARC | 8010–8028 |
| Reverse | AYYAAGCCTCCTACTAYYATTATGAA | 8277–8302 |
| Forward | TGGGGHTGCTCTGGAARAC | 8010–8028 |
| Reverse | AYYAARCCTCCTAYTAYCATTATGAA | 8277–8302 |
| Probe | NED-CARCARGAAAWRAATGAA-MGB | 8178–8195 |
| Alternative probe | NED-TGGGANARAGAAATT-MGB | 8115–8129 |
Underlined characters in the inner primers indicate signature nucleotide positions that can discriminate between human immunodeficiency virus type 1 (HIV-1) subtype B and CRF01_AE.
Multiplex real-time PCR.
The HIV-1 RNA was then reverse transcribed into cDNA using SuperScript III RNase H Reverse Transcriptase (Invitrogen, Carlsbad, CA) and random hexamers (Applied Biosystems, Carlsbad, CA). The first round PCR mix contained MyFi Mix, 2× (Bioline, UK), 400 nM of each outer primer, and 3 μL of cDNA. Nuclease-free water was added up to a final volume of 50 μL per reaction. The thermal cycling profile was as follows: initial denaturation at 95°C for 3 min, 30 cycles of 98°C for 20 s, 44°C for 1 min and 72°C for 1 min, and final extension at 72°C for 10 min in a DNA Engine Tetrad2 Thermal Cycler (BioRad, Hercules, CA).
The real-time PCR mix in the second round PCR contained 12.5 μL of SensiFAST Probe Lo-ROX (Bioline), 400 nM of each primer set of inner primer (except 800 nM of forward primer for the pro-RT region), 200 nM of probe for p24 region, 300 nM for pro-RT region, and 400 nM for gp41 region in a final volume of 25 μL. The TaqMan probes were labeled with different fluorescent reporter dyes at 5′-end and each probe selectively hybridized to the specific target region. The probes within the p24, pro-RT, and gp41 regions were labeled with the FAM, VIC, and NED dyes (Applied Biosystems), respectively. All probes were labeled with non-fluorescent quencher minor groove binder at the 3′-end. The real-time amplification PCR was performed on the ViiA 7 Real-Time PCR System (Applied Biosystems) with the following thermal profiles: 95°C for 2 min, 40 cycles of 97°C for 15 s, and 55°C for 1 min 30 s. Of note, we have included viral isolates with confirmed HIV-1 genotypes (subtype B, CRF01_AE, and CRF33_01B) previously determined in our laboratory as in-house controls for every run.
A classification system based on the real-time PCR results or yield of the targeted regions were used to interpret the HIV-1 subtype or CRF in the samples (Table 2). A quantification cycle (Cq) rule25 derived based on the targeted regions was applied to improve the accuracy for the differential identification of HIV-1 subtypes/CRFs. The Cq value was determined by the lowest take-off point above the background noise of the amplification curve. The optimum difference between the Cq values (ΔCq) for the p24, pro-RT, and gp41 regions of the same sample was determined. The Cq values of more than 38 cycles were disregarded. It is also essential to note that both CRF01_AE and CRF53_01B shared similar real-time PCR yield and were not readily distinguished by the assay (Table 2). Genetic sequencing followed by phylogenetic analysis (refer below) were therefore performed for precise genotype classification.
Classification of HIV-1 genotypes based on the real-time PCR amplification and detection of the p24, pro-RT, and gp41 genes
| HIV-1 genotype | p24 (CRF01_AE-specific) | Primers and probes | gp41 (subtype B-specific) |
|---|---|---|---|
| pro-RT (subtype B-specific) | |||
| Subtype B | − | + | + |
| CRF01_AE | + | − | − |
| CRF33_01B | + | + | − |
| CRF53_01B* | + | − | − |
| CRF54_01B | − | + | − |
| CRF58_01B | + | + | + |
CRF01_AE and CRF53_01B shared similar real-time PCR yield.
“+” and “−” signs indicate positive and negative real-time PCR detection, respectively.
HIV-1 = human immunodeficiency virus type 1.
A panel of calibrated standards of HIV-1 subtypes obtained from the NIH AIDS Research and Reference Reagent Program was used for assay validation and the determination of limit of detection (LOD). The panel included HIV-1 subtype B (US1, BK132, and BZ167) and CRF01_AE (CM235, CM240, and IN12), which were originated from the United States, Brazil, Thailand, and Indonesia.26 Each sample was run in duplicates. The real-time PCR results were then analyzed and the genotype was determined based on the protocol described previously. A 10-fold serial dilution of standards with known viral load was used to assess the LOD. The HIV-1 negative plasma specimens (N = 50) were also used to determine the specificity of the assay.
To validate our method, we tested a different set of field samples collected among fishermen from a rural community in Kuantan (more than 200 km east of Kuala Lumpur) was conducted. A total of 410 subjects with histories of injecting drug use were recruited in 2011. The HIV-1 screening was performed using the ACON HIV Rapid Test Kit (ACON Laboratories, San Diego, CA) and positive results were confirmed with the Intec HIV Rapid Test Kit (Intec Products Inc., Xiamen, China). Plasma samples from HIV-positive subjects were transported to the laboratory in Kuala Lumpur for real-time PCR analysis.
Sequencing and phylogenetic analysis.
For accurate HIV-1 genotype determination, the conventional direct Sanger sequencing approach and phylogenetic analysis were performed. Nested PCR was performed on each sample using the in-house primers described previously to amplify the gag-pol gene (HXB2: 625–3440nt).8 Sequencing reaction was carried out in an ABI PRISM 3730xl Genetic Analyzer using the BigDye Terminator v3.1 cycle sequencing kit chemistry (Applied Biosystems, Foster City, CA). The nucleotide sequences were edited manually and the gag-pol contigs were aligned with the HIV-1 reference subtypes and CRFs downloaded from the LANL HIV sequence database. The reference strains of HIV-1 genotypes used in the alignment were CRF01_AE, subtype B, CRF33_01B, CRF48_01B, CRF51_01B, CRF53_01B, CRF54_01B, and CRF58_01B. Neighbor-joining phylogenetic analysis was carried out using the Kimura 2-parameter model in MEGA version 6.0.27 The Recombinant Identification Program (RIP) available at the LANL HIV sequence database and bootscanning analysis were used for the characterization of recombinant mosaic genomes.28 The TaqMan real-time PCR assay results were then compared with the phylogenetic data for HIV-1 genotype confirmation.
Results
Assay development and validation.
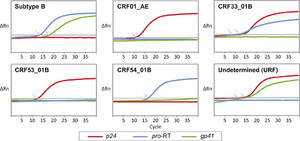
A nested multiplex real-time PCR assay was developed to detect HIV-1 subtype B, CRF01_AE, CRF33_01B, and newly emerging genotypes circulating in Southeast Asia. Primers and probes of the assay targeting multiple genetic regions was designed using global reference sequences of Asian origin based on the genome organizations of the circulating subtypes and CRFs in the region. The performance of the assay was evaluated on 180 HIV-positive clinical samples (heterosexual, 31.1%; male homosexual, 22.2%; bisexual, 3.9%; injecting drug user [IDU], 10.6%; unreported risk, 32.2%). The median viral load for these samples was 6.7 log10 copies/mL (range: 3.0–7.0 log10 copies/mL). The real-time PCR results of the targeted regions (gag p24, pol pro-RT, and env gp41 regions) were used to interpret and determine the HIV-1 subtype or CRF (representative amplification curves are shown in Figure 2), based on the classification chart in Table 2 and the previously mentioned ΔCq threshold, which was determined to be 5.6 cycles. Genotype assignment was then confirmed by direct sequencing and phylogenetic analysis. Five distinct HIV-1 genotypes were detected: subtype B (16.7%, 30 of 180), CRF01_AE (52.8%, 95 of 180), CRF33_01B (24.4%, 44 of 180), CRF53_01B (1.1%, 2 of 180), and CRF54_01B (0.6%, 1 of 180) (Table 3). Of note, real-time PCR results from eight specimens showed discordant genotype assignment when compared with nucleotide sequence analysis. These specimens were genotyped by real-time PCR as CRF01_AE (N = 3), CRF33_01B (N = 3), CRF58_01B (N = 1), and an “undetermined” (p24+, pro-RT, gp41+) genotype (N = 1). Recombination analysis of the gag-pol segments using the RIP program and bootscanning showed that these isolates were unique recombinant sequences involving CRF01_AE and subtype B, termed as CRF01_AE/B URFs (4.4%, 8 of 180). Only one specimen showed real-time PCR results (as CRF01_AE) that were not concordant with the phylogenetic inference (as CRF33_01B), as a result of the probe mismatch in the pro-RT region (Table 3). The panel of HIV-1 subtype B (US1, BK132, and BZ167) and CRF01_AE (CM235, CM240, and IN12) were successfully genotyped by the assay under different dilutions (108 to 103 copies/mL), with no evidence of cross-priming.



Real-time PCR amplification plot for human immunodeficiency virus type 1 (HIV-1) genotypes. Representative amplification curves of the p24, pro-RT, and gp41 genes for subtype B, CRF01_AE, CRF33_01B, CRF53_01B, CRF54_01B, and an undetermined genotype from Kuala Lumpur were shown. The quantification cycle (Cq) value was indicated by an arrow. CRF01_AE, CRF33_01B, and CRF53_01B showed positive amplification in the p24 region (red); subtype B, CRF33_01B, and CRF54_01B in the pro-RT (blue); and subtype B in the gp41 region (green).
Citation: The American Society of Tropical Medicine and Hygiene 92, 3; 10.4269/ajtmh.14-0681
HIV-1 genotype determination by real-time PCR assay and phylogenetic analysis
| HIV-1 genotype by phylogenetic analysis | HIV-1 genotype by real-time PCR | ||||||
|---|---|---|---|---|---|---|---|
| Subtype B | CRF01_AE / CRF53_01B | CRF33_01B | CRF54_01B | CRF58_01B | Undetermined | Total | |
| Subtype B | 30 | 30 | |||||
| CRF01_AE | 95 | 95 | |||||
| CRF33_01B | 1 | 43 | 44 | ||||
| CRF53_01B | 2 | 2 | |||||
| CRF54_01B | 1 | 1 | |||||
| CRF58_01B | 0 | ||||||
| CRF01_AE/B URF | 3 | 3 | 1 | 1 | 8 | ||
| Total | 30 | 101 | 46 | 1 | 1 | 1 | 180 |
HIV-1 = human immunodeficiency virus type 1; URF = unique recombinant form.
Overall, the accuracy of the real-time PCR assay to correctly identify an HIV-1 genotype was 95.0% (171 of 180), in which the sensitivities for subtype B, CRF01_AE, and CRF33_01B classification were 100.0% (30 of 30), 100.0% (95 of 95, 7% (43 of 44), respectively. Inter-genotype specificity was more than 95% in general, in which the specificities for subtype B, CRF01_AE, and CRF33_01B were calculated at 100.0% (150 of 150), 95.2% (79 of 83), and 97.8% (133 of 136), respectively. The assay also showed high specificity in HIV-negative specimens (100%, 50 of 50). The positive and negative predictive values of the assay for the major subtypes were both 100.0% for subtype B, 96.0% and 100.0% for CRF01_AE, and 93.5% and 99.3% for CRF33_01B, respectively. These values for other rare CRFs were not calculated as a result of inadequate sample size. The limit of detection of the assay was determined at 103 copies/mL for both subtype B and CRF01_AE.
In the field test, a total of 36 subjects (8.8%) tested positive for HIV-1 infection among 410 drug injecting fishermen recruited from a rural village in Kuantan, Pahang. Real-time PCR results, in combination with sequence and phylogenetic analysis, showed the presence of HIV-1 subtype B, CRF01_AE, CRF33_01B, and CRF01_AE/B URF at 8.3% (3 of 36), 11.1% (4 of 36), 66.7% (24 of 36), and 2.8% (1 of 36), respectively. Four samples were incorrectly genotyped by the assay because of primer/probe mismatches. The overall accuracy of the assay was 86.1% (31 of 36).
Discussion
In this study, a nested real-time PCR assay capable of detecting the common and newly emerging HIV-1 subtypes and CRFs circulating in Southeast Asia was designed and evaluated. The prevalent or endemic HIV-1 genotypes targeted by the assay were subtype B, CRF01_AE, and CRF33_01B, whereas the recently described and relatively rare recombinant lineages were CRF53_01B, CRF54_01B, and CRF58_01B. The conceptual design of the assay was based on the mosaic genome structures of the various recombinant lineages and the genetic signatures within the p24, pro-RT, and gp41 regions that could distinguish between subtype B and CRF01_AE (Figure 1). Genotype-specific primers and probes were then designed to target either subtype B or CRF01_AE-related segments, enabling the differential detection of six HIV-1 genotypes based on the real-time PCR yield in these genetic regions (Table 2).
The newly developed genotyping assay was highly sensitive and specific, accurately determined more than 90% of the circulating HIV-1 genotypes in Kuala Lumpur, Malaysia. The results were in concordance with sequence and phylogenetic analysis for genotype determination, underlining the usefulness of the assay as an alternative method to provide rapid genotyping results, particularly in studies where the sample sizes are large. The performance of the assay was also comparable with other similar genotyping assays previously developed.19–23 It is however important to note that, in addition to the common or classical circulating genotypes, our assay is capable of detecting novel recombinant lineages (CRF53_01B, CRF54_01B, and CRF58_01B) recently reported in Malaysia.10,11,13 Likewise, the multi-region approach of the assay allows the detection of HIV-1 URFs in a time-effective manner,22 a technically established strategy for tracing the emergence of yet undefined CRF lineages in the region.
From 180 specimens collected in Kuala Lumpur between 2008 and 2012 among subjects who acquired HIV infection mainly through sexual contacts, the real-time PCR assay showed that more than half of the population were infected with CRF01_AE, followed by CRF33_01B (Table 3). The molecular epidemiological data determined by the newly developed genotyping tool was largely in concordance with previous studies that reported the predominance of HIV-1 CRF01_AE among the sexual population during the 2003–2010 period.6,29 Although the results suggest a consistent distribution of genetic diversity in the last decade, testing of recently collected samples showed the presence of novel recombinant genotypes such as CRF53_01B and CRF54_01B, albeit at low frequencies. The availability of a rapid and versatile genotyping assay will therefore facilitate the tracking of unique or complex genotypes that are otherwise difficult to detect via phylogenetic analysis based on limited HIV-1 genome coverage.
In a field test conducted among HIV-positive drug injecting fishermen from a rural village, the TaqMan assay showed high sensitivity in determining the HIV-1 genotypes. Unlike the urban population in Kuala Lumpur, HIV-1 CRF33_01B was detected at high prevalence, followed distantly by subtype B and CRF01_AE. Although both subtype B and CRF01_AE were the predominant genotypes during the early phase of the epidemic in the region,2 recent studies reported significant genotype replacement in which CRF33_01B became the major circulating genotype among the IDUs.8,12 The widespread dissemination of CRF33_01B among IDUs further reinforced the epidemiological and public health impact attributed to emerging recombinant lineages in the region.
The performance of the genotyping assay, however, could be affected by nucleotide mismatches in the primer and/or probe binding regions. As indicated in Table 1, the specificity of the assay is mainly dependent on single nucleotide modification near the 3′-end region of the primer. Amplification may occur in samples where mismatches were present in both the forward and reverse primers, producing false positive fluorescent signals that can be detected at the late phase of the amplification cycles. In addition, similar to other multi-region PCR assay, amplification efficiency may vary between different genetic regions in the genome.22 Given the limited potential sites for primer design, particularly for a highly diverse virus such as the HIV-1, such a problem is difficult to avoid in assay design. Therefore, to enhance the power of differential identification for various HIV-1 subtypes and CRFs, the difference in Cq values between the target genes was used to assess the PCR efficiency.25 Another possible factor that could affect the performance of the assay is the presence of mixed infections by multiple HIV-1 genotypes. For example, if a specimen contains both subtype B and CRF01_AE, the real-time PCR assay will probably show positive detection for all three target regions (p24+, pro-RT+, gp41+), in which case the sample will be misclassified as CRF58_01B according to Table 2. Clonal sequencing and phylogenetic analysis may be required for precise genotype classification. Cases of mixed infections, however, were not apparent in this study. Finally, although the real-time PCR assay is capable of detecting unique recombinant lineages, such descriptions can only be limited to the genetic regions targeted by the assay. Inter-genotype recombination event(s) that occur outside the p24, pro-RT, and gp41 regions could be missed by the assay and as a result, the degree of viral genetic diversity or complexity will be underestimated as a result of mis-classification. Expanding the assay coverage beyond these regions will certainly improve the resolution for URF detection,22 but the proportionate increase in cost and labor may restrict the use of the assay in resource-limited settings. In any case, if the cost for genetic sequencing is not the limiting factor, a genotyping method that targets more genetic regions or the complete viral genome will be useful for accurate and reliable genotype classification.
In summary, the newly developed genotyping assay is accurate, simple to perform, and suitable for genotype determination in areas where the genetic diversity of HIV-1 is high.30 The availability of a sensitive real-time PCR assay that can detect a panel of circulating genotypes simultaneously may be useful for continuous molecular epidemiological surveillance, particularly in the Southeast Asia region.
ACKNOWLEDGMENTS
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: US1, BK132, BZ167, CM235, CM240, and IN12 from Nelson Michael. We thank Nur Ezreen Syafina, Yeat Mei Lee, Siti Humaira Razak, Nur Afiq Idham, and Hisyammudin Nawi for technical assistance.
- 2.↑
Ou CY, Takebe Y, Weniger BG, Luo CC, Kalish ML, Auwanit W, Yamazaki S, Gayle HD, Young NL, Schochetman G, 1993. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet 341: 1171–1174.
- 3.↑
Chen JH, Wong KH, Chen Z, Chan K, Lam HY, To SW, Cheng VC, Yuen KY, Yam WC, 2010. Increased genetic diversity of HIV-1 circulating in Hong Kong. PLoS ONE 5: e12198.
- 4.
Lee CC, Sun YJ, Barkham T, Leo YS, 2009. Primary drug resistance and transmission analysis of HIV-1 in acute and recent drug-naive seroconverters in Singapore. HIV Med 10: 370–377.
- 5.
Sahbandar IN, Takahashi K, Djoerban Z, Firmansyah I, Naganawa S, Motomura K, Sato H, Kitamura K, Pohan HT, Sato S, 2009. Current HIV type 1 molecular epidemiology profile and identification of unique recombinant forms in Jakarta, Indonesia. AIDS Res Hum Retroviruses 25: 637–646.
- 6.↑
Tee KK, Li XJ, Nohtomi K, Ng KP, Kamarulzaman A, Takebe Y, 2006. Identification of a novel circulating recombinant form (CRF33_01B) disseminating widely among various risk populations in Kuala Lumpur, Malaysia. J Acquir Immune Defic Syndr 43: 523–529.
- 7.↑
van Hal SJ, Herring B, Deris Z, Wang B, Saksena NK, Dwyer DE, 2009. HIV-1 integrase polymorphisms are associated with prior antiretroviral drug exposure. Retrovirology 6: 12.
- 8.↑
Chow WZ, Ong LY, Razak SH, Lee YM, Ng KT, Yong YK, Azmel A, Takebe Y, Al-Darraji HA, Kamarulzaman A, Tee KK, 2013. Molecular diversity of HIV-1 among people who inject drugs in Kuala Lumpur, Malaysia: Massive expansion of circulating recombinant form (CRF) 33_01B and emergence of multiple unique recombinant clusters. PLoS ONE 8: e62560.
- 9.↑
Robertson DL, Anderson JP, Bradac JA, Carr JK, Foley B, Funkhouser RK, Gao F, Hahn BH, Kalish ML, Kuiken C, Learn GH, Leitner T, McCutchan F, Osmanov S, Peeters M, Pieniazek D, Salminen M, Sharp PM, Wolinsky S, Korber B, 2000. HIV-1 nomenclature proposal. Science 288: 55–56.
- 10.↑
Chow WZ, Al-Darraji H, Lee YM, Takebe Y, Kamarulzaman A, Tee KK, 2012. Genome sequences of a novel HIV-1 CRF53_01B identified in Malaysia. J Virol 86: 11398–11399.
- 11.↑
Chow WZ, Takebe Y, Syafina NE, Prakasa MS, Chan KG, Al-Darraji HA, Koh C, Kamarulzaman A, Tee KK, 2014. A newly emerging HIV-1 recombinant lineage (CRF58_01B) disseminating among people who inject drugs in Malaysia. PLoS ONE 9: e85250.
- 12.↑
Li Y, Tee KK, Liao H, Hase S, Uenishi R, Li XJ, Tsuchiura T, Yang R, Govindasamy S, Yong YK, Tan HY, Pybus OG, Kamarulzaman A, Takebe Y, 2010. Identification of a novel second-generation circulating recombinant form (CRF48_01B) in Malaysia: a descendant of the previously identified CRF33_01B. J Acquir Immune Defic Syndr 54: 129–136.
- 13.↑
Ng KT, Ong LY, Takebe Y, Kamarulzaman A, Tee KK, 2012. Genome sequence of a novel HIV-1 circulating recombinant form 54_01B from Malaysia. J Virol 86: 11405–11406.
- 14.↑
Liu Y, Li L, Bao Z, Li H, Zhuang D, Liu S, Wang X, Li T, Jia L, Yang S, Li J, 2012. Identification of a novel HIV type 1 circulating recombinant form (CRF52_01B) in Southeast Asia. AIDS Res Hum Retroviruses 28: 1357–1361.
- 15.
Tovanabutra S, Kijak GH, Beyrer C, Gammon-Richardson C, Sakkhachornphop S, Vongchak T, Jittiwutikarn J, Razak MH, Sanders-Buell E, Robb ML, Suriyanon V, Birx DL, Michael NL, Celentano DD, McCutchan FE, 2007. Identification of CRF34_01B, a second circulating recombinant form unrelated to and more complex than CRF15_01B, among injecting drug users in northern Thailand. AIDS Res Hum Retroviruses 23: 829–833.
- 16.↑
Tovanabutra S, Watanaveeradej V, Viputtikul K, De Souza M, Razak MH, Suriyanon V, Jittiwutikarn J, Sriplienchan S, Nitayaphan S, Benenson MW, Sirisopana N, Renzullo PO, Brown AE, Robb ML, Beyrer C, Celentano DD, McNeil JG, Birx DL, Carr JK, McCutchan FE, 2003. A new circulating recombinant form, CRF15_01B, reinforces the linkage between IDU and heterosexual epidemics in Thailand. AIDS Res Hum Retroviruses 19: 561–567.
- 17.↑
Ng OT, Eyzaguirre LM, Carr JK, Chew KK, Lin L, Chua A, Leo YS, Redd AD, Quinn TC, Laeyendecker O, 2012. Identification of new CRF51_01B in Singapore using full genome analysis of three HIV type 1 isolates. AIDS Res Hum Retroviruses 28: 527–530.
- 18.↑
Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, Yao JD, Wengenack NL, Rosenblatt JE, Cockerill FR 3rd, Smith TF, 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev 19: 165–256.
- 19.↑
Freitas FB, Esteves A, Piedade J, Parreira R, 2013. Novel multiregion hybridization assay for the identification of the most prevalent genetic forms of the human immunodeficiency virus type 1 circulating in Portugal. AIDS Res Hum Retroviruses 29: 318–328.
- 20.
Hoelscher M, Dowling WE, Sanders-Buell E, Carr JK, Harris ME, Thomschke A, Robb ML, Birx DL, McCutchan FE, 2002. Detection of HIV-1 subtypes, recombinants, and dual infections in east Africa by a multi-region hybridization assay. AIDS 16: 2055–2064.
- 21.
Kijak GH, Sanders-Buell E, Wolfe ND, Mpoudi-Ngole E, Kim B, Brown B, Robb ML, Birx DL, Burke DS, Carr JK, McCutchan FE, 2004. Development and application of a high-throughput HIV type 1 genotyping assay to identify CRF02_AG in West/West Central Africa. AIDS Res Hum Retroviruses 20: 521–530.
- 22.↑
Kijak GH, Tovanabutra S, Sanders-Buell E, Watanaveeradej V, de Souza MS, Nelson KE, Ketsararat V, Gulgolgarn V, Wera-arpachai M, Sriplienchan S, Khamboonrueng C, Birx DL, Robb ML, McCutchan FE, 2007. Distinguishing molecular forms of HIV-1 in Asia with a high-throughput, fluorescent genotyping assay, MHAbce v.2. Virology 358: 178–191.
- 23.↑
Wei M, Guan Q, Liang H, Chen J, Chen Z, Hei F, Feng Y, Hong K, Huang H, Xing H, Shao Y, 2004. Simple subtyping assay for human immunodeficiency virus type 1 subtypes B, C, CRF01-AE, CRF07-BC, and CRF08-BC. J Clin Microbiol 42: 4261–4267.
- 24.↑
Katoh K, Asimenos G, Toh H, 2009. Multiple alignment of DNA sequences with MAFFT. Methods Mol Biol 537: 39–64.
- 25.↑
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT, 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622.
- 26.↑
Jagodzinski LL, Wiggins DL, McManis JL, Emery S, Overbaugh J, Robb M, Bodrug S, Michael NL, 2000. Use of calibrated viral load standards for group M subtypes of human immunodeficiency virus type 1 to assess the performance of viral RNA quantitation tests. J Clin Microbiol 38: 1247–1249.
- 27.↑
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S, 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729.
- 28.↑
Salminen MO, Carr JK, Burke DS, McCutchan FE, 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res Hum Retroviruses 11: 1423–1425.
- 29.↑
Ong LY, Razak SN, Lee YM, Sri La Sri Ponnampalavanar S, Syed Omar SF, Azwa RI, Tee KK, Kamarulzaman A, 2014. Molecular diversity of HIV-1 and surveillance of transmitted drug resistance variants among treatment Naive patients, 5 years after active introduction of HAART in Kuala Lumpur, Malaysia. J Med Virol 86: 38–44.
- 30.↑
Hemelaar J, Gouws E, Ghys PD, Osmanov S, 2011. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS 25: 679–689.