INTRODUCTION
Dracunculiasis, or Guinea worm (GW) disease, is targeted for global eradication by the World Health Assembly.1 The disease is caused by the roundworm Dracunculus medinensis. A person becomes infected after drinking water containing small crustaceans (copepods) that are themselves infected with the third-stage larvae of D. medinensis. After a 10- to 14-month incubation period, the gravid adult female worm, measuring up to 1 m in length, emerges through a blister that forms on the skin, most often on the lower leg or foot. The blister is associated with a burning pain that frequently drives the infected person to seek relief by immersing the affected body part in cool water. When the worm detects the temperature difference on submersion, it discharges first-stage larvae into the water, and the larvae are ingested by copepods where they undergo two molts to become infectious third-stage larvae and begin the life cycle again.2
The ongoing efforts of the GW Eradication Program (GWEP) are now focused on six countries in Africa currently affected by GW (Angola, Cameroon, Chad, Ethiopia, Mali, and South Sudan).3–6 In 2019, 54 human GW cases were reported from Chad, South Sudan, Angola, and Cameroon (likely imported from Chad).7 As the human case count eventually trends toward zero, identification of each and every case will become increasingly important for the global elimination of the parasite. Guinea worm disease is diagnosed in the field by its clinical presentation—emergence of a string-like worm from a skin lesion. With the decline in human cases reported globally, all human cases should be laboratory confirmed at the U.S. CDC by protocol since 2014.8 This involves sending a piece of the emerged worm in alcohol to the CDC for detailed evaluation including morphologic examination and PCR assay.9,10 In those instances where rumors of individuals with GW disease symptoms are reported to health officials but no worm specimen is collected or available for laboratory verification, a sensitive and specific serologic assay would be useful as means of case confirmation or exclusion. Serologic assays may also have some use in focal surveillance of communities where outbreaks of unknown magnitude are ongoing, especially if the assay is shown to detect specific antibody responses during the asymptomatic, prepatent period of the GW infection.
In addition, recombinant GW antigens identified in these human studies may have future use in species-specific assays for the assessment of population-level infection dynamics in dogs, cats, and baboons that have recently become a concern as reservoirs for GW transmission in some parts of sub-Saharan east Africa. Sustained GW transmission in dogs has been ongoing in Chad since 2012.11 Since 2014, dog cases have also been reported in Ethiopia and Mali, although in far fewer numbers than in Chad. Single dog cases have also been reported in South Sudan (2015) and Angola (2019). Guinea worm cases in cats (Chad, Ethiopia, and Mali) and baboons (Ethiopia) have been reported as well.9,12,13 Genome sequencing, microsatellite genotype analysis, and mitochondrial haplotype evaluation have confirmed that the human and animal GW specimens are all D. medinensis and are genetically indistinguishable.11,13,14 Research on the mechanisms of animal transmission is now pointing toward the possible ingestion of transport hosts, such as fingerling fish or fish entrails containing infected copepods and/or paratenic hosts such as frogs.15–17 Human behavior is likely playing a role in GW transmission to dogs and cats. For example, in Chad, mass fish harvesting occurs at the end of the dry season along the country’s largest river system, the Chari River and its tributaries, and this period coincides with the peak in monthly incidence of GW cases in dogs (May–June). Humans may facilitate animal access to small fish and fish guts.11,18
Throughout the history of the GWEP, there has been no commercially available or field-ready laboratory test for humans or animals to diagnose infection with D. medinensis. Research on the immunologic responses of humans to D. medinensis infections was conducted in sample populations in Nigeria (late 1980s),19 Ghana (1989–1991),20–23 and Togo (2005)24 with future aspirations toward possible development of a serologic assay that might detect prepatent, patent, and post-patent infections. Although these early studies suggested that D. medinensis is immunogenic, they relied on crude worm antigen preparations and failed to identify the specific antigens responsible for the immunogenicity and antibody recognition. They did, however, note that IgG and, specifically, the subclasses of IgG1 and IgG4 were elevated in the prepatent, patent, and post-patent periods (P < 0.05).22,24 Cross-reactivity observed with sera from patients with Onchocerca volvulus and other helminth infections was recognized as a possible challenge to further test development.19,20,23
Since these early studies, laboratory methods have evolved. In this study, we took advantage of a set of residual human plasma samples from dracunculiasis-infected donors and of the availability of excellent and well-characterized antihuman IgG secondary antibody reagents to identify immunodominant GW antigen targets. We then developed serologic multiplex antibody assays for anti-GW antibodies in human samples using recombinant forms of two of the newly identified protein antigens. Future validation of these antibody assays using dog and nonhuman primate sera could provide a new and valuable tool for use in the final stages of the international GW disease eradication program.
MATERIALS AND METHODS
Study samples.
Anonymized, residual human plasma samples from a previously reported GW study in Togo in 2005 were available for analysis.24 The plasma set included five samples from donors with patent GW infection, 19 samples from donors who reported previous GW infection (collected 1–8 years after worm emergence), and 12 samples from donors who never reported GW infection. The plasma samples were diluted with an equal volume of RPMI 1640 (Gibco, Eggenstein, Germany) medium at the time of collection and were stored frozen. CDC staff had no contact with donors and no access to personal identifying information. The participants in the original study in Togo (those with patent and post-patent GW infections and those selected as endemic controls) provided informed consent to use their samples in GW assays to study antibody reactivity in body fluids, including tears, serum, and plasma. The Ministry of Health of Togo provided authorization for the original study.17
For specificity determinations, the following anonymous serum samples were available for testing: 25 sera from skin snip–positive donors with O. volvulus infection from the Democratic Republic of the Congo, Ethiopia, or Uganda; 10 sera from microfilaremic donors with Wuchereria bancrofti from India or Haiti; five sera from donors infected with Loa loa from Cameroon; five sera from donors infected with Mansonella ozzardi from Peru; and 49 sera from Argentina residents with stool-confirmed Strongyloides stercoralis infections with or without other soil-transmitted helminths. The specificity sample set also included eight anonymous sera from healthy adult blood donors from a region of Haiti that was highly endemic for W. bancrofti.
Residual GW adult female worm segments from diagnostic human specimens were used with CDC human subjects’ approval.
Antigen preparation.
An ∼2-cm segment of GW collected from a dog and stored in ethanol was dried at room temperature under vacuum (< 2 mm Hg) in a SpeedVac concentrator (Savant Instruments, Farmingdale, NY) for at least 4 hours. The segment was cut into 1 mm pieces and suspended in 0.5 mL of buffer containing 50 mM tris(hydroxymethyl)-aminomethane at pH 7.5, 2 mM ethylenediaminetetraacetic acid (EDTA), 100 mM NaCl, and 1% sodium dodecyl sulfate (SDS). The pieces were ground with a pestle, heated to 95°C for 10 minutes, and sonicated for 30 seconds (Heat Systems, model W-255 sonicator with a microtip at setting number 4) (Heat Systems-Ultrasonics, Farmingdale, NY). After a 10-minute centrifugation (3,000 × g) at 4°C to remove insoluble material, the protein concentration of the supernatant was determined using the bicinchoninic acid (BCA) microassay with bovine serum albumin as the standard (Pierce, Rockford, IL). The protein yield from the SDS extraction procedure was approximately 4 mg. An equal volume of 2× SDS loading buffer with 20% β-mercaptoethanol was added, and the sample was heated at 95°C for 5 minutes before electrophoresis.25
Alternatively, an ∼2 cm segment of dried worm from a human patient was cut into 1 mm pieces and suspended in 0.5 mL of buffer containing 50 mM Na2HPO4 at pH 7.5, 100 mM NaCl, 2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), 0.2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, and 2 mM N-ethylmaleimide. The worm was ground with a pestle at room temperature, and 50 μL of Triton X-114 (TX-114) was then added with further grinding at 4°C. The suspended worm pieces were sonicated as described earlier and then subjected to a freeze-thaw cycle. Following the removal of insoluble worm material by centrifugation at 4°C (10 minutes, 3,000 × g), TX-114 phase partition extraction was performed using the method of Ko and Thompson.26,27 Proteins were precipitated from the detergent-enriched phase and the detergent-depleted aqueous phase (Aq) with the addition of 4 volumes of acetone and overnight storage at −20°C. The precipitates were collected by centrifugation (10 minutes, 3,000 × g, 4°C), dried at room temperature, resuspended in 0.5 mL of phosphate-buffered saline (PBS; 10 mM Na2HPO4 at pH 7.2 and 0.85% NaCl) with 1% SDS, and heated at 95°C for 5 minutes. The protein yield in the Aq was variable depending on the quality of the worm segment but was often in the range of 300–450 μg. An equal volume of 2× SDS loading buffer with 20% β-mercaptoethanol was added before polyacrylamide gel electrophoresis.25
Sodium dodecyl sulfate polyacrylamide gels and Western blot analysis.
Total SDS-extracted GW proteins (1.5–2 μg/mm in a 130-mm preparative well) were resolved on 10–22.5% gradient SDS polyacrylamide gels using the Laemmli buffer system25 and then electro-blotted onto polyvinylidene difluoride membrane (PVDF; Immobilon P, Millipore Corporation, Bedford, MA). Pre-stained protein markers were included on each polyacrylamide gel to monitor electrophoretic progress (ThermoFisher Scientific, Waltham, MA). The blotted membrane was cut into 2-mm strips, and strips were incubated overnight at 4°C in 2 mL of 1:200 dilutions of each test plasma in 0.3% Tween-20 in PBS. Bound total IgG antibodies were detected using dilutions of biotinylated mouse antihuman IgG (clone H2 from Southern Biotech, Birmingham, AL) and biotinylated mouse antihuman IgG4 (clone HP6025 from Southern Biotech) together in the same incubation. Bound IgG subclass antibodies were detected using dilutions of biotinylated mouse antihuman IgG1 (clone HP6069 from Invitrogen, Camarillo, CA), antihuman IgG2 (clone HP6002 from Invitrogen), antihuman IgG3 (clone HP6047 from Invitrogen), or antihuman IgG4 (clone HP6025 from Southern Biotech). Secondary antibodies and streptavidin/alkaline phosphatase were diluted 1:1,000 in 0.3% Tween-20 PBS, and each was incubated with the strips for 1 hour at room temperature. Blots were developed as previously described.27 AuroDye Forte colloidal gold stain (GE Healthcare, Piscataway, NJ) was used to visualize the total protein profile on a blot strip.
To determine whether the antigens were composed of protein or carbohydrate, on-blot digestion was performed using the method of Priest et al.28 Polyvinylidene difluoride strips of SDS-extracted GW proteins were incubated with either 25 mM NaIO4 in 50 mM sodium acetate buffer at pH 4.5 (overnight at room temperature) or with 100 μg/mL proteinase K in PBS buffer (overnight at 37°C). After extensive washing of the periodate-treated strips with 1% glycine in PBS and extensive washing of the proteinase-treated strips with PBS containing 0.3% Tween-20 and 1 mM PMSF, Western blots were conducted for either total IgG or IgG4 subclass responses as described earlier using a human plasma with a strong positive GW response.
Antigen sequencing and identification.
Triton X-114 extraction Aq proteins (approximately 300 μg in a 130-mm preparative well or 65 μg in a 35-mm preparative well) were resolved by SDS polyacrylamide gel electrophoresis as described earlier. The gels were stained overnight in a solution of 0.5 mg/mL Coomassie brilliant blue R-250 (Sigma-Aldrich Co., St. Louis, MO) in 7% acetic acid and 50% methanol. The gel was destained in a solution of 15% acetic acid and 10% methanol until protein bands were visible. The protein band corresponding to Western blot–reactive antigen was excised from the gel along with equivalent sized segments of gel immediately above and below the protein band. All three gel slices were washed with 50% acetonitrile, washed with 100% acetonitrile, and then stored at 4°C for peptide sequencing.
Gel slices were cubed with a scalpel into 1 mm pieces and washed three times with 50% acetonitrile and 10 mM (NH4)HCO3. The samples were dried in a SpeedVac rotary evaporator, reduced with dithiothreitol in 10 mM (NH4)HCO3 at 37°C for 1 hour followed by alkylation with iodoacetamide in 10 mM (NH4)HCO3 at 25°C for 1 hour. Gel samples were washed with 10 mM (NH4)HCO3 followed by two washes with 50% acetonitrile and 10 mM (NH4)HCO3. Samples were dried in a SpeedVac and reconstituted with trypsin (V5111, Promega Corporation, Madison, WI) in 10 mM (NH4)HCO3. Digests were incubated at 37°C overnight. Tryptic digests were extracted using two washes of 60% acetonitrile and 10 mM (NH4)HCO3, and were dried in a SpeedVac. Samples were submitted for nano-liquid chromatography- mass spectrometry/mass spectrometry (nano-LC-MS/MS) analysis.
Nano-LC-MS/MS mass spectrometric analysis was performed using a ThermoFisher Scientific model Lumos Electrospray Ionization tribrid orbitrap instrument interfaced with an on-line EASY-Spray nanospray source (ThermoFisher Scientific) to perform LC-MS/MS using a RSLCnano HPLC (ThermoFisher Scientific) configured for nanoliter per-minute flows. A desalting trap column (0.3 × 5 mm, 5 μm C18 PepMap 120 A, ThermoFisher Scientific) was used, and the analytical column used was a C18 EASY-Spray column (0.075 × 250 mm, 2 μm, 120A, ThermoFisher Scientific) operated at a column temperature of 35°C. The solvents used were 0.1% formic acid in water (A) and 80% acetonitrile/0.1% formic acid (B). The gradient was 2–35% B in 120 minutes. The source was operated at a spray voltage of 1900V. The capillary temperature was set to 275°C. The mass spectrometer was set to data-dependent acquisition mode with a 3-second cycle time. A single MS spectrum was acquired in the orbitrap followed by as many MS/MS spectra as could be acquired in the ion trap during the 3-second cycle. The MS spectra were acquired in the orbitrap using an m/z range of 350–1,500 at a resolution of 120,000 with a maximum injection time of 50 ms and an automatic gain control (AGC) target of 200,000 ions. The MS/MS data were acquired in the linear ion trap with an automated m/z range starting at 110 m/z using the “Rapid” scan mode with a maximum ion inject time of 300 ms and an AGC target of 3,000 ion using collision-induced dissociation fragmentation at 35% collision energy and quadrupole isolation of 1.6 Da. A precursor was selected based on intensity and charge state (charges between 2 and 7), and precursor exclusion was set for 20 seconds after one spectrum was acquired for each parent ion.
The collected data were processed by Proteome Discoverer (v. 2.2.0.388, ThermoFisher Scientific) using the built-in Sequest HT search engine. The data were searched against the D. medinensis protein database downloaded from the WormBase public database (www.wormbase.org)29,30 (accessed April 2, 2018 and July 18, 2018) with added common contaminating proteins such as human keratins. False discovery rates (FDRs) and quality scores (q-value, post error probability) were calculated for each peptide and protein hit using the Percolator node in Proteome Discoverer. “High” confidence proteins and peptides used a FDR cutoff of 0.01, and “medium” confidence proteins and peptides used a cutoff of 0.05.
Antigen cloning, expression, and analysis.
Dracunculus medinensis protein-coding sequences of interest were optimized for bacterial expression and synthesized by GenScript as inserts between the BamHI and EcoRI restriction enzyme sites of pGEX4T-2 plasmid (GE Healthcare). Plasmids were transformed into Escherichia coli HB101 cells as directed by the manufacturer (Promega Corporation). Schistosoma japonicum glutathione-S-transferase (GST) fusion proteins were expressed in isopropyl-β-D-thiogalactopyranoside–induced bacterial cultures and purified by glutathione Sepharose 4B column chromatography as directed by the manufacturer (GST bulk purification module; GE Healthcare). Glutathione-S-transferase protein was expressed from pGEX4T-2 plasmid with no DNA insert and purified as described for use as a negative control.31 Purified protein preparations were dialyzed overnight at 4°C against > 300 volumes of PBS buffer (SpectraPor3 membrane, 3,500-Da cutoff; Spectrum Laboratories, Rancho Dominguez, CA) and assayed for protein content using the BCA microassay (Pierce). Purified GST fusion proteins (approximately 6 μg/well) were resolved on 12% polyacrylamide mini gels using the method of Laemmli.25 One gel was stained with Coomassie Brilliant Blue R-250 (Bio-Rad, Hercules, CA), whereas another was electro-blotted onto PVDF membrane. A Western blot was performed using a pool of 1:200 plasma from GW-infected patients, a 1:1,000 mouse antihuman IgG4, and a 1:1,000 streptavidin/alkaline phosphatase as described earlier.
The GW domain of unknown function 148 (DUF148) and thioredoxin-like protein 1 (TRXL1)–GST fusion proteins were digested overnight at room temperature with thrombin as directed by the manufacturer (GE Healthcare) and then passed over a glutathione Sepharose 4B column to remove the released GST and uncleaved fusion protein. The protein concentration of the flow-through fraction containing the purified antigen of interest with no GST was determined by BCA microassay. Purified TRXL1 and DUF148 antigens without GST (approximately 6 μg/well) along with approximately 10 μg of TX-114 Aq GW antigen were resolved on a 10–22.5% gradient SDS polyacrylamide gel using the Laemmli buffer system25 and then transferred onto a PVDF membrane. A Western blot was performed using a pool of 1:200 plasma from GW-infected patients, a 1:1,000 mouse antihuman IgG4 dilution, and a 1:1,000 streptavidin/alkaline phosphatase dilution as described earlier.
Bead coupling and multiplex bead assays.
Protocols for coupling of antigens to SeroMap beads (Luminex Corporation, Austin, TX) have recently been described in detail by Priest and Moss.32 For the current study, the GST control protein was coupled at a concentration of 15 μg protein/12.5 × 106 beads in 25 mM 2-(N-morpholino)-ethanesulfonic acid buffer with 0.85% NaCl at pH 5.0. Domain of unknown function 148–GST, TRXL1–GST, heat shock protein 1 (HSP1)–GST, and HSP2–GST proteins were each coupled using 120 μg protein/12.5 × 106 beads in PBS buffer at pH 7.2.
Sera were diluted 1:400 in buffer containing PBS at pH 7.2, 0.5% polyvinyl alcohol, 0.8% polyvinylpyrrolidone, 0.5% casein, 0.3% Tween-20, 0.02% NaN3, and 3 μg/ml E. coli extract.32,33 For the plasma samples from Togo, the 1:1 pre-dilution in the RPMI medium was considered in the final 1:400 dilution calculation. The conditions previously described by Priest and Moss32 for serum incubation, secondary antibody binding, streptavidin-coupled R-phycoerythrin binding, and final buffer wash were used for the GW multiplex bead assays. Total IgG antibody assays used 1 μg/mL biotinylated mouse antihuman IgG (clone H2; Southern Biotech) plus 0.8 μg/mL biotinylated mouse antihuman IgG4 (clone HP6025; Southern Biotech) as the detection reagent and were run in duplicate.32 Multiplex bead assays for IgG4 antibody detection used only the biotinylated mouse antihuman IgG4 antibody (0.8 μg/mL) and were run in single wells. Assays were read using a Bio-Plex 200 instrument with Bio-Plex Manager version 6.2 software, build 175 (Bio-Rad). Results for single-well IgG4 assays are expressed as the median fluorescent intensity (MFI) without a blank correction. Average results for duplicate well IgG assays are expressed as the MFI value minus the value for the buffer-only background blank (MFI-bg). If ≥ 2 positive responses from a test sample had coefficients of variation > 15% between the duplicate wells, then the assay was repeated.
Data analysis.
Mature protein-coding sequences were predicted using SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP).34 Sequences were compared using the basic local alignment search tool for proteins from the National Center for Biotechnology Information.35,36 Cutoff values were determined either by Youden’s J-index or by receiver-operating characteristic (ROC) curve analysis with the closest to (0,1) criterion.37–39 Receiver-operating characteristic curves were generated, and statistical analyses (the Kruskal–Wallis one-way analysis of variance and the Mann–Whitney rank-sum test) were conducted using SigmaPlot 13.0 (Systat Software, Inc., San Jose, CA). An alpha level of 0.05 was used to determine statistical significance.
RESULTS
Identification of immunodominant GW antigens.
In a preliminary experiment, Western blots for total IgG antibodies using plasma from GW disease and endemic normal donors from Togo (Supplemental Figure 1) did not reveal clearly identifiable and well-isolated antigen bands that could be used to differentiate between infected and uninfected individuals. An IgG subclass analysis using the plasma from one GW disease patient demonstrated that the IgG response was dominated by IgG4 antibodies with a less intense IgG1 component (Supplemental Figure 2). Western blots for IgG4 antibodies using the Togo plasma panel demonstrated a clear pattern of antigen recognition on blot strips incubated with GW disease donor plasma that was largely absent on blot strips incubated with endemic normal plasma (Figure 1). A total of 15 of 24 plasma samples (62%) from GW-disease donors had detectable (although sometimes quite faint) IgG4 antibody reactivity to antigens in the 10 to −11 kDa size range, whereas none of the endemic negative donors had similar reactivity.



IgG4 Western blots of plasma samples from GW-infected and GW-uninfected individuals from Togo. Guinea worm proteins extracted into SDS buffer as described in Materials and Methods were resolved on 10–22.5% gradient SDS polyacrylamide gels (1.5–2 μg/mm in a 130-mm preparative well) and then electro-blotted onto PVDF membrane. Membrane strips (2 mm) were incubated in 1:200 dilutions of each test plasma in 0.3% Tween-20 in PBS, and bound IgG4 antibodies were detected using a biotinylated monoclonal mouse antihuman IgG4 antibody and the streptavidin/alkaline phosphatase system. B indicates a control strip incubated with Tween-PBS buffer alone. Au indicates a strip incubated with AuroDye Forte to stain all bound proteins. The apparent molecular weights of See Blue Plus 2 pre-stained markers (ThermoFisher Scientific) are indicated on the left. The location of the antigen band of interest is indicated by an arrow. GW = Guinea worm; MWapp = apparent molecular weights; PBS = phosphate-buffered saline; PVDF = polyvinylidene difluoride; SDS = sodium dodecyl sulfate.
Citation: The American Journal of Tropical Medicine and Hygiene 103, 6; 10.4269/ajtmh.20-0511
To determine whether the epitopes of interest were protein or carbohydrate in nature, strips of GW antigen were treated overnight with either sodium periodate to cleave the vicinal diols of carbohydrates or with proteinase K to digest the peptide bonds of proteins. As shown in Supplemental Figure 3, proteinase K digestion eliminated antibody binding to the blot strip, whereas sodium periodate treatment had no effect on either IgG or IgG4 antibody recognition of the antigen band.
The antigen of interest was enriched in the detergent-depleted, aqueous fraction following TX-114 extraction of dried GW fragments (Figure 2). Surprisingly few proteins were extracted into the TX-114 detergent-enriched fraction, and most of the proteins remained in the insoluble pellet. This may reflect the fact that the worm had been submerged in ethanol for an extended period of time for transport and for storage before extraction.



TX-114 phase-114 extraction of antigens from GW. Antigens from the detergent-depleted Aq, the detergent-enriched TX-114 phase (Tx), and the insoluble GW pellet (Insol) obtained as described in the Materials and Methods were resolved on a 10–22.5% gradient SDS polyacrylamide gel and then electro-blotted onto the PVDF membrane. One panel was stained with AuroDye Forte colloidal gold (left), whereas the other panel (right) was incubated with a 1:200 pool of plasma from GW-positive donors and developed to visualize bound IgG4 antibodies as described in the Materials and Methods. The apparent molecular weights of See Blue Plus 2 pre-stained markers (ThermoFisher Scientific) are indicated on the left. The location of the antigen band of interest is indicated by an arrow. Aq = aqueous phase; GW = Guinea worm; MWapp = apparent molecular weights; PVDF = polyvinylidene difluoride; SDS = sodium dodecyl sulfate; TX-114 = Triton X-114.
Citation: The American Journal of Tropical Medicine and Hygiene 103, 6; 10.4269/ajtmh.20-0511
Protein sequencing, cloning, and expression.
Potential protein sequences of interest were identified from two separate peptide mass sequencing runs of TX-114-extracted Aq GW proteins. Even though the target antigen migrated on SDS polyacrylamide gels with an apparent molecular weight significantly below 16 kDa, an upper molecular weight limit of 18 kDa was chosen for the search criteria to account for potential posttranslational modifications such as signal peptide cleavage. Eight candidate proteins were identified based on the predicted molecular weights and the relative distributions of the proteins between the excised gel band and the two control gel segments cut from the regions immediately above and below the band (Table 1). The potential proteins of interest included TRXL1 and TRXL2, HSP1 and HSP2, two transthyretin-like proteins (THY1 and THY2), a DUF148, and an unknown protein sequence. Predicted mature protein molecular weights were determined from the coding sequences using SignalP-5.0 (Table 1).34 Basic local alignment search tool for proteins comparison of the predicted D. medinensis protein sequences to predicted proteins encoded in other nematode worm species’ genomes revealed a large number of highly conserved homologues. For example, the D. medinensis DUF148 sequence exhibits 44% identity at the amino acid level to the Toxocara canis homologue (Supplemental Figure 4A), and the D. medinensis TRXL1 sequence exhibits 49% identity to the T. canis thioredoxin domain–containing protein 12 homologue (Supplemental Figure 4B).40
Accession numbers and identities of potential proteins of interest identified by mass sequencing of excised polyacrylamide gel bands containing Guinea worm proteins
| Accession number | Identity | Abbreviation | Predicted CDS MW (kDa) | Mature protein MW (kDa)* |
|---|---|---|---|---|
| DME_0000774101-mRNA-1 | Thioredoxin-like protein | TRXL1 | 17.8 | 15.9 |
| DME_0000113301-mRNA-1 | Domain of unknown function 148 | DUF148 | 16.0 | 14.2 |
| DME_0000286601-mRNA-1 | Transthyretin-like protein | THY1 | 15.7 | 13.3 |
| DME_0000083201-mRNA-1 | Heat shock protein | HSP1 | 12.8 | 12.8 |
| DME_0000929301-mRNA-1 | Thioredoxin-like protein | TRXL2 | 12.8 | 12.8 |
| DME_0000801701-mRNA-1 | Heat shock protein | HSP2 | 12.7 | 12.7 |
| DME_0000148801-mRNA-1 | Transthyretin-like protein | THY2 | 12.7 | 10.4 |
| DME_0000028101-mRNA-1 | Unknown protein | SMU1 | 12.3 | 10.4 |
MW = molecular weight.
Predicted using SignalP 5.0 to identify cleaved amino-terminal signal peptide sequence.
Predicted mature protein coding sequences for all eight of the proteins of interest were synthesized and inserted into plasmids for expression as GST fusion proteins. Six of the proteins were expressed in the soluble fraction in E. coli and could be purified by glutathione Sepharose 4B column chromatography. Transthyretin-like protein1 and THY2 expression products were found in the inclusion bodies and were not further studied. When the soluble, purified GST fusion proteins were resolved on a 12% SDS polyacrylamide gel (Figure 3A), transferred to PVDF membrane, and exposed to a pool of human GW infection plasma for a Western blot, IgG4 antibody reactivity was noted to bands corresponding to the DUF148, TRXL1, HSP1, and HSP2 proteins (Figure 3B). Domain of unknown function 148 and TRXL1 mature proteins, cleaved from their respective GST fusion proteins by thrombin treatment and purified by glutathione Sepharose 4B column chromatography, were run on a 10–22.5% gradient SDS polyacrylamide gel alongside a sample of TX-114-extracted Aq GW proteins. After transfer to PVDF membrane and exposure to a pool of human GW infection plasma for a Western blot, IgG4 antibody reactivity to the DUF148 and TRXL1 proteins was noted at approximately the same molecular weight as the target band in the GW aqueous protein extract lane (indicated in Supplemental Figure 5 by an arrow).



Analysis of purified recombinant GW GST fusion proteins. Soluble, recombinant GW proteins purified by glutathione Sepharose 4B column chromatography were resolved together on one 12% SDS polyacrylamide gel as described in the Materials and Methods. The gel in Panel A was stained with Coomassie Brilliant Blue R-250. Another gel was electro-blotted onto the PVDF membrane, incubated with a 1:200 pool of plasma from GW-positive donors, and developed to visualize bound IgG4 antibodies as described in the Materials and Methods (Panel B). Lane designations are as follows: pre-stained molecular weight standards, S; DUF148, D; TRXL1, T1; HSP1, H1; HSP2, H2; SMU1, U; and TRXL2, T2. The apparent molecular weights of See Blue Plus 2 pre-stained markers (ThermoFisher Scientific) are indicated on the right, and the apparent molecular weights of PageRuler Plus pre-stained markers (ThermoFisher Scientific) are indicated on the left. DUF = domain of unknown function; GW = Guinea worm; GST = glutathione-S-transferase; HSP = heat shock protein; MWapp = apparent molecular weights; PVDF = polyvinylidene difluoride; SDS = sodium dodecyl sulfate; SMU = unknown protein sequence; TRXL = thioredoxin-like protein; TX-114 = Triton X-114.
Citation: The American Journal of Tropical Medicine and Hygiene 103, 6; 10.4269/ajtmh.20-0511
Multiplex bead assays.
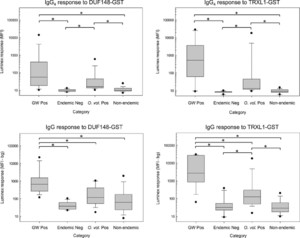
IgG4 and total IgG antibody responses to recombinant GW fusion proteins were assessed by multiplex bead assay (Figure 4). Plasma samples from residents of Togo who had current or a previous history of GW infection at the time of sample donation had IgG4 antibody responses to DUF148–GST (median = 56 MFI; range = 10–16,534) and to TRXL1–GST (median = 518 MFI; range = 9–29,222) that were significantly higher than responses observed using plasma from endemic normal individuals from Togo or non-endemic sera. For both IgG4 assays, responses to sera from O. volvulus–positive donors were significantly higher than responses from the endemic and non-endemic sample sets. Using the total IgG antibody assay, plasma samples from GW-positive donors had responses to DUF148–GST (median = 654.5 MFI-bg; range = 143–24,271) and to TRXL1–GST (median = 2,735 MFI-bg; range = 58–30,933) that were significantly higher than responses observed using plasma from endemic normal individuals from Togo, sera from O. volvulus–positive donors, or non-endemic sera. Onchocerca volvulus–positive donor serum responses were significantly higher than responses from the endemic and non-endemic sample sets for total IgG TRXL1–GST assay.



Multiplex bead assay detection of IgG4 and IgG antibodies to recombinant DUF148–GST and TRXL1–GST fusion proteins. Multiplex bead assays were conducted as described in the Materials and Methods using SeroMap beads covalently coated with either DUF148–GST (left panels) or TRXL1–GST (right panels). Appropriate biotinylated monoclonal secondary antibodies were used to detect either IgG4 (top panels) or total IgG (bottom panels). Antibody response distributions for the GW-positive plasma samples from Togo (GW pos, n = 24), plasma samples from uninfected controls from Togo (Endemic Neg, n = 12), sera from patients with confirmed infection of Onchocerca volvulus (O. volvulus Pos., n = 25), and sera from non-endemic regions with other nematode infections (non-endemic, n = 77) are represented by box plots. Boxes include values between the 25th and 75th percentiles. Whiskers and data points represent the 10th and 90th and the 5th and 95th percentiles, respectively. Median values are indicated by a horizontal line within the boxes. Distributions that show statistically significant differences (P < 0.05) using the Kruskal–Wallis one-way analysis of variance on ranks with pairwise multiple comparison procedures are indicated by brackets with asterisks. DUF = domain of unknown function; GST = glutathione-S-transferase; GW = Guinea worm; TRXL = thioredoxin-like protein.
Citation: The American Journal of Tropical Medicine and Hygiene 103, 6; 10.4269/ajtmh.20-0511
In contrast to the TRXL1–GST and DUF148–GST assays described earlier, only low IgG4 and total IgG responses were observed to the recombinant HSP1–GST (median = 10 MFI and 17 MFI-bg, respectively) and HSP2–GST (median = 11 MFI and 18 MFI-bg, respectively) proteins using plasma from GW-positive donors. Heat shock protein 1–GST and HSP2–GST responses for GW-positive donor plasma and responses for endemic negative donors from Togo were not significantly different using the Mann–Whitney rank-sum test (HSP1–GST IgG4 P-value = 0.72; HSP2–GST IgG4 P-value = 0.80; HSP1–GST IgG P-value = 0.40; HSP2–GST IgG P-value = 0.18). Given the low HSP responses among the GW-positive donors, these antigens were not included in multiplex assay testing of the O. volvulus–positive and non-endemic serum sets. Beads coupled with GST protein, included in each assay well as a negative control, had responses ranging from 19 to 39 MFI (median = 24; mean = 25) for IgG4 assays and from −2 to 100 MFI-bg (median = 4.5; mean = 7) for total IgG assays.
Receiver-operating characteristic curves using results from the complete set of test samples (24 positive plasma samples from GW-infected donors and 114 negative samples from individuals who never had GW) were used to identify cutoff values for the IgG4 multiplex bead assays (Supplemental Figure 6). For the DUF148-GST and TRXL1-GST IgG4 multiplex assays, cutoffs of 17.5 MFI and 19.5 MFI units were identified, respectively, but the value of 17.5 MFI units for the DUF148-GST assay was not above the limit of detection as determined from the three plate blank values (mean + 3 SDs = 22 MFI). The summary of the IgG4 responses for the various categories of sample donors is given in Table 2. The DUF148-GST multiplex assay had a sensitivity of 79.2% and a specificity of 85.1%, with an area under the ROC curve of 0.86 (Supplemental Figure 6). The TRXL1-GST multiplex assay had a sensitivity of 87.5% and a specificity of 93.9%, with an area under the curve of 0.90. If a “simultaneously positive by both assays” definition was applied to the IgG4 antibody response data set, then the sensitivity would be 79.2%, and the overall specificity of the test would improve to 94.7%, with only 20% of O. volvulus donors classified as positive (Table 2).
Human serum IgG antibody reactivity to Guinea worm antigens
| Donor group | Number | DUF148 IgG4, n (%) | TRXL1 IgG4, n (%) | Both* IgG4, n (%) | DUF148 IgG, n (%) | TRXL1 IgG, n (%) | Both* IgG, n (%) | DUF148 IgG HC,† n (%) | TRXL1 IgG HC,† n (%) | Both* IgG HC,† n (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| D. medinensis, patent | 5 | 3 (60)‡ | 4 (80) | 3 (60) | 4 (80) | 5 (100) | 4 (80) | 4 (80) | 5 (100) | 4 (80) |
| D. medinensis, post-patent ≤ 1 year | 4 | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) | 4 (100) |
| D. medinensis, post-patent > 1 year | 15 | 12 (80) | 13 (87) | 12 (80) | 15 (100) | 13 (87) | 13 (87) | NU§ | NU§ | NU§ |
| Onchocerca volvulus | 25 | 12 (48) | 6 (24) | 5 (20) | 9 (36) | 7 (28) | 3 (12) | 9 (36) | 4 (16) | 2 (8) |
| Wuchereria bancrofti | 10 | 2 (20) | 1 (10) | 1 (10) | 2 (20) | 0 | 0 | 1 (10) | 0 | 0 |
| Loa loa | 5 | 1 (20) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mansonella ozzardi | 5 | 1 (20) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Strongyloides stercoralis | 49 | 0 | 0 | 0 | 15 (31) | 3 (6) | 1 (2) | 10 (20) | 2 (4) | 1 (2) |
| Haiti non-endemic | 8 | 1 (12) | 0 | 0 | 3 (37) | 0 | 0 | 3 (37) | 0 | 0 |
| Togo endemic negative | 12 | 0 | 0 | 0 | 0 | 1 (8) | 0 | 0 | 0 | 0 |
D. medinensis = Dracunculus medinensis; DUF148 = domain of unknown function 148; GST = glutathione-S-transferase; ROC = receiver-operating characteristic; MFI-bg = median fluorescent intensity minus background; TRXL1 = thioredoxin-like protein 1. Summary of multiplex bead assay results using ROC assigned cutoffs for DUF148–GST and TRXL1–GST coated beads. Except for the last column of this table, cutoffs were defined using ROC curve analysis of the entire sample set of 24 positives and 114 negatives. For the last column, only the nine samples from patent and recent infection donors were used in the ROC analysis.
Simultaneously positive for responses to both DUF148–GST and TRXL1–GST antigens.
HC: high cutoffs of 199.5 and 754 MFI-bg, respectively, were used for the DUF148–GST and TRXL1–GST assay results reported in these columns.
Number positive (%).
NU: not used in the ROC curve analysis.
Areas under the ROC curves were slightly higher for the DUF148-GST (area under the curve = 0.88) and TRXL1-GST (area under the curve = 0.95) total IgG multiplex assays (Supplemental Figure 7) than for the IgG4 assays described earlier. Using a ROC-identified cutoff of 161.5 MFI-bg, the DUF148-GST multiplex assay was 95.8% sensitive and 74.6% specific with 36% of the responses of O. volvulus donors classified as positive (Table 2). Using a ROC-identified cutoff of 236.5 MFI-bg, the TRXL1-GST multiplex assay was 91.7% sensitive and 90.4% specific with 28% of the responses of O. volvulus donors classified as positive. Both cutoffs were greater than the respective limits of quantitation (37 MFI; 27 MFI). If, however, a “simultaneously positive by both assays” definition was applied to the antibody response data set, then the overall specificity of the test would improve to 96.5% with only 12% of O. volvulus donors classified as positive (Table 2). Although the sensitivity of the combined antigen test would decrease to 87.5%, Youden’s J-index suggests that the combined test is superior to either individual assay (0.84 versus 0.70 and 0.82).
Domain of unknown function 148-GST and TRXL1-GST IgG antibody responses were higher in samples collected during GW patency or within 1 year of worm emergence (n = 9) than in samples collected more than 1 year after illness resolution (n = 15) (Figure 5), and the observed difference was statistically significant in the case of the TRXL1-GST assay (Mann–Whitney rank-sum test; P = 0.007). If only responses from this currently infected/recently recovered patient category (≤ 1 year after worm emergence) were used as the positive set in the ROC analysis, the area under the ROC curve would improve to 0.99 (Figure 6). At a ROC-defined cutoff of 754 MFI-bg, the sensitivity of the TRXL1-GST IgG assay was 100% with a specificity of 94.7%, and only 16% of the responses of O. volvulus donors were classified as positive (Table 2). Using a “simultaneously positive by both assays” definition with a DUF148-GST IgG assay cutoff of 199.5 MFI-bg would improve overall specificity to 97.4% but would decrease the sensitivity to 88.9%, and Youden’s J-index for the combined test (J-index = 0.86) would be lower than that of the TRXL1-GST assay alone (J-index = 0.95) because of the misclassification of one positive sample.



The magnitude of the IgG response to recombinant GW proteins varies with time since worm emergence. Box plots show the distributions of total IgG responses for DUF148-GST and TRXL1-GST multiplex bead assays of plasma samples from GW-positive donors from Togo. Responses of samples (n = 9) collected from individuals with patent infection or from individuals within 1 year of worm emergence (≤ 1) are plotted separately from the responses of samples (n = 15) collected from individuals more than 1 year after worm emergence (> 1). Box plot characteristics are as described in Figure 4. Distributions that show statistically significant differences (P < 0.05) using the Mann–Whitney rank-sum test are indicated by a bracket with an asterisk. DUF = domain of unknown function; GST = glutathione-S-transferase; GW = Guinea worm; TRXL = thioredoxin-like protein.
Citation: The American Journal of Tropical Medicine and Hygiene 103, 6; 10.4269/ajtmh.20-0511



Receiver-operating characteristic curve results using total IgG responses from DUF148–GST (black line) and TRXL1–GST (red line) multiplex bead assays. Curves were constructed using MFI-bg responses of samples (n = 9) collected from individuals with a patent GW infection or from individuals within 1 year of GW emergence and responses of samples (n = 114) collected from individuals who never had GW. The optimal threshold for the DUF148–GST assay was 199.5 MFI-bg units with a sensitivity of 88.9% and a specificity of 79.8%. The optimal threshold for the TRXL1–GST assay was 754 MFI-bg units with a sensitivity of 100% and a specificity of 94.7%. DUF = domain of unknown function; GW = Guinea worm; GST = glutathione-S-transferase; MFI-bg = median fluorescent intensity minus background; ROC = receiver-operating characteristic; TRXL = thioredoxin-like protein.
Citation: The American Journal of Tropical Medicine and Hygiene 103, 6; 10.4269/ajtmh.20-0511
DISCUSSION
The first successful attempt at the development of a serologic assay for GW disease used first-stage larvae as antigen in a total IgG indirect fluorescence assay (IFA).41 This IFA by Muller41 had a sensitivity of 97% using sera from individuals with patent infections, and the antibody titers waned in the months following worm emergence. Muller41 was also the first to document GW assay cross-reactivity with sera from onchocerciasis patients (17% cross-reactivity by IFA). He conducted experimental infections in rhesus monkeys and demonstrated that detectable antibody titers appeared several months before worm emergence.41 Other groups have used soluble antigen fractions extracted from adult female worms or first-stage larvae to develop ELISA, dot ELISA, and Falcon assay screening test ELISA methods that are easier to use and more amenable to large-scale surveys.19,20,23,24,42 The ELISA results confirmed that antibody responses increase in the prepatent period and decline following resolution of the infection.19,21–23 Although ELISA specificity was often improved by focusing on the IgG4 antibody response, cross-reactivity with sera from onchocerciasis patients was consistently observed.19,20,23,43 Multiple laboratories have previously observed that GW assay specificity was higher when using a low molecular weight fraction of soluble GW antigen (12–20 kDa size range) than when using a fraction containing larger size antigens.19,20,43 We did, in fact, observe some higher molecular weight antigens by the IgG4 Western blot that appeared to react with sera from individuals who never reported GW infection. One of these antigens, a galectin with a molecular weight of 30 kDa, reacted extensively with sera from donors who never had GW (data not shown).
In this work, we report the development of a multiplex bead assay based on two low molecular weight antigens identified from an aqueous extract of adult female D. medinensis worms. The 14.2 kDa GW DUF148 antigen is member of the SXP-1 family of proteins that includes the Brugia malayi Bm14 antigen44,45 and the O. volvulus Ov17 antigen.46,47 SXP-1 antigens are characterized by strong IgG responses that are often skewed to the IgG4 subclass and by considerable antibody cross-reactivity between antigens from related parasitic nematode species of humans.44,47–49 Consistent with these reports, cross-reactivity with sera from O. volvulus–positive donors was high with both DUF148-GST multiplex assays (IgG4 = 48%; IgG = 36%). Bloch and Simonsen reported that the distribution of IgG subclass responses to GW antigens varied between individuals.20 With our assay, we also see some individuals who have responses heavily skewed to the IgG4 subclass as well as some individuals who lack IgG4 responses altogether. Despite a 10% loss of specificity, the much-improved sensitivity of the IgG assay compared with the IgG4 assay (95.8% versus 79.2%, respectively) resulted in a higher J-index for the DUF148-GST IgG assay.
The second low molecular weight antigen identified by our peptide sequencing efforts and used for multiplex assay, the 15.9 kDa GW TRXL1, is a member of the thioredoxin superfamily of redox proteins that contain a CysXaaYaaCys motif.50 Thioredoxins and TRXL have been identified in parasitic nematodes as components of the excretory/secretory fraction and are believed to be involved in host immunomodulation.51,52 As with the GW DUF148-GST assay described earlier, cross-reactivity with sera from onchocerciasis patients was observed with the TRXL1-GST multiplex assays (IgG4 = 24%; IgG = 28%), but the overall specificities of both assays were > 90%. The sensitivities and specificities of the TRXL1-GST IgG and IgG4 assays differed by only about 4% such that the J-indices of both assays were comparable. Based on J-index comparisons and ROC areas, the TRXL1-GST assays were superior to the DUF148-GST assays.
Bloch and Simonsen suggested that a combination of two or more assay results representing different classes or subclasses of antibodies might produce an improved overall method for the detection of GW infection.20 Although we could not simultaneously measure more than one type of antibody response in a single assay well, the multiplex bead assay format does allow for the measurement of a single type of antibody response (total IgG or IgG4, in our case) to multiple antigens in one assay well. When we reevaluated the results using a “simultaneously positive for antibodies to two antigens” definition, we observed high specificities for both the IgG4 and IgG assays (94.7% and 96.5%, respectively). Given that antibodies to GW antigens decline with time after worm emergence,19,21–23 we excluded results from plasma collected > 1 year after GW emergence and repeated the ROC curve analysis for the IgG assay. The higher cutoffs assigned by this analysis resulted in the highest specificity of any assay we measured (97.4% when using the “positive-for-two” definition), with only two (8%) of the O. volvulus–positive and only one (2%) of the strongyloidiasis-positive donors misclassified. We would note that an added advantage of the multiplex bead assay format is that highly specific markers for O. volvulus, B. malayi, and S. stercoralis infection could easily be incorporated into the method to highlight those individuals who might be at increased risk of misclassification due to potential antibody cross-reactivity from non-GW nematode infections.47,53,54 With a sensitivity of 88.9%, the high-cutoff, “positive-for-two” total IgG assay J-index (0.86) is comparable to that of the IgG4, low-molecular-weight GW larval antigen ELISA reported by Bloch and Simonsen (0.88).20
The success of the GWEP in minimizing the number of human cases of GW disease worldwide has resulted in one of the major weaknesses of this work: namely, that we only had plasma collected in 2005 from 24 GW-positive donors in Togo for analysis, and only nine of these were collected during or immediately after worm emergence. A misclassification of only one response from among these nine samples would have a major impact on the overall interpretation of assay performance. Additional studies of human samples collected during or immediately after GW emergence would improve confidence in the quality of the assay. Alternatively, the assays could be further tested after modification for use with sera from dogs, cats, or baboons, as infections in those animals are now more common than infections in humans. Likely, all that is required is a specific, low-background secondary antibody against the IgGs of each species. A second weakness of our study is that we lacked prepatent samples from individuals who were infected with GW. For the assay to be an effective tool for serosurveillance, we need to demonstrate that it can detect specific antibody responses before the worm becomes detectable. Current large-scale studies of dogs in highly endemic regions of Chad could provide the sample set necessary to validate the assay in prepatent populations if a functional dog assay can be developed. Finally, our GW-positive samples came from only one small location in Togo that represents only a small part of the original parasite geographic range. Although it is possible that antigen sequence differences between isolated populations might impact sensitivity, this scenario is unlikely, given recent DNA sequence analysis of GW populations.13,14
In conclusion, we report the development of a new multiplex bead assay using two recombinant, low molecular weight GW antigens. The IgG4 and total IgG TRXL1-GST assays both performed well with sensitivities > 87% and specificities > 90%. Maximum specificity (> 96%) was obtained with the total IgG assay when both the TRXL1-GST and DUF148-GST tests were used to define a positive case. We are now working to adapt the assay for use with other animal species.
ACKNOWLEDGMENTS
We would like to thank Alejandro Krolewiecki for providing serum samples from strongyloidiasis-positive donors and Mark Eberhard for assistance in selection of GW segments. We thank Alan Tracey and James Cotton of the Wellcome Sanger Institute for access to an updated Dracunculus medinensis genome annotation and for checking the accuracy of the gene models for our target antigens. We would also like to thank the participants of the original study in Togo, the Ministry of Health of Togo, and the researchers at the Institut National d’Hygiène at the Programme Nationale de Lutte contre la Dracunculose in Lomé, Togo, the Université de Lomé at the Faculté Mixte de Médecine et Pharmacie in Lomé, Togo, and the Institute for Tropical Medicine at the University of Tübingen in Germany, who provided the residual plasma samples and without whom this work would not have been possible.
REFERENCES
- 2.↑
Ruiz-Tiben E, Hopkins DR, 2006. Dracunculiasis (Guinea worm disease) eradication. Adv Parasitol 61: 275–309.
- 3.↑
Hopkins DR, Ruiz-Tiben E, Eberhard ML, Roy SL, Weiss AJ, 2016. Progress toward global eradication of dracunculiasis-January 2015–June 2016. MMWR Morb Mortal Wkly Rep 65: 1112–1116.
- 4.
Hopkins DR, Ruiz-Tiben E, Eberhard ML, Roy SL, Weiss AJ, 2017. Progress toward global eradication of dracunculiasis, January 2016–June 2017. MMWR Morb Mortal Wkly Rep 66: 1327–1331.
- 5.
Hopkins DR, Ruiz-Tiben E, Weiss AJ, Roy SL, Zingeser J, Guagliardo SAJ, 2018. Progress toward global eradication of dracunculiasis–January 2017–June 2018. MMWR Morb Mortal Wkly Rep 67: 1265–1270.
- 6.↑
Hopkins DR, Weiss AJ, Roy SL, Zingeser J, Guagliardo SAJ, 2019. Progress toward global eradication of dracunculiasis–January 2018–June 2019. MMWR Morb Mortal Wkly Rep 68: 979–984.
- 7.↑
WHO Collaborating Center for Dracunculiasis Eradication, CDC, 2020. Guinea Worm Wrap-Up #266. Available at: https://www.cartercenter.org/resources/pdfs/news/health_publications/guinea_worm/wrap-up/266.pdf. Accessed April 24, 2020.
- 8.↑
WHO Collaborating Center for Dracunculiasis Eradication, CDC, 2014. Guinea Worm Wrap-Up #228. Available at: https://www.cartercenter.org/resources/pdfs/news/health_publications/guinea_worm/wrap-up/228.pdf. Accessed April 24, 2020.
- 9.↑
World Health Organization, 2017. Dracunculiasis eradication: global surveillance summary, 2016. Wkly Epidemiol Rec 92: 269–286.
- 10.↑
Bimi L, Freeman AR, Eberhard ML, Ruiz-Tiben E, Pieniazek NJ, 2005. Differentiating Dracunculus medinensis from D. insignis, by the sequence analysis of the 18S rRNA gene. Ann Trop Med Parasitol 99: 511–517.
- 11.↑
Eberhard ML et al. 2014. The peculiar epidemiology of dracunculiasis in Chad. Am J Trop Med Hyg 90: 61–70.
- 12.↑
WHO Collaborating Center for Dracunculiasis Eradication, CDC, 2019. Guinea Worm Wrap-Up #261. Available at: https://www.cartercenter.org/resources/pdfs/news/health_publications/guinea_worm/wrap-up/261.pdf. Accessed September 26, 2019.
- 13.↑
Thiele EA, Eberhard ML, Cotton JA, Durrant C, Berg J, Hamm K, Ruiz-Tiben E, 2018. Population genetic analysis of Chadian Guinea worms reveals that human and non-human hosts share common parasite populations. PLoS Negl Trop Dis 12: e0006747.
- 14.↑
Durrant C et al. 2019. Population genomic evidence that human and animal infections in Africa come from the same populations of Dracunculus medinensis. BioRxiv. Availble at: https://doi.org/10.1101/808923.
- 15.↑
Cleveland CA, Eberhard ML, Thompson AT, Garrett KB, Swanepoel L, Zirimwabagabo H, Moundai T, Ouakou PT, Ruiz-Tiben E, Yabsley MJ, 2019. A search for tiny dragons (Dracunculus medinensis third-stage larvae) in aquatic animals in Chad, Africa. Sci Rep 9: 375.
- 16.
Cleveland CA, Eberhard ML, Thompson AT, Smith SJ, Zirimwabagabo H, Bringolf R, Yabsley MJ, 2017. Possible role of fish as transport hosts for Dracunculus spp. larvae. Emerg Infect Dis 23: 1590–1592.
- 17.↑
Eberhard ML, Yabsley MJ, Zirimwabagabo H, Bishop H, Cleveland CA, Maerz JC, Bringolf R, Ruiz-Tiben E, 2016. Possible role of fish and frogs as paratenic hosts of Dracunculus medinensis, Chad. Emerg Infect Dis 22: 1428–1430.
- 18.↑
WHO Collaborating Center for Research TaEoD, CDC, 2014. Guinea Worm Wrap-Up #230. Available at: https://www.cartercenter.org/resources/pdfs/news/health_publications/guinea_worm/wrap-up/230.pdf. Accessed September 26, 2019.
- 19.↑
Fagbemi BO, Hillyer GV, 1990. Immunodiagnosis of dracunculiasis by Falcon assay screening test-enzyme-linked immunosorbent assay (FAST-ELISA) and by enzyme-linked immunoelectrotransfer blot (EITB) technique. Am J Trop Med Hyg 43: 665–668.
- 20.↑
Bloch P, Simonsen PE, 1998. Studies on immunodiagnosis of dracunculiasis. I. Detection of specific serum antibodies. Acta Trop 70: 73–86.
- 21.↑
Bloch P, Simonsen PE, 1998. Immunoepidemiology of Dracunculus medinensis infections I. Antibody responses in relation to infection status. Am J Trop Med Hyg 59: 978–984.
- 22.↑
Bloch P, Simonsen PE, 1998. Immunoepidemiology of Dracunculus medinensis infections II. Variation in antibody responses in relation to transmission season and patency. Am J Trop Med Hyg 59: 985–990.
- 23.↑
Bloch P, Simonsen PE, Vennervald BJ, 1993. The antibody response to Dracunculus medinensis in an endemic human population of northern Ghana. J Helminthol 67: 37–48.
- 24.↑
Knopp S, Amegbo IK, Hamm DM, Schulz-Key H, Banla M, Soboslay PT, 2008. Antibody and cytokine responses in Dracunculus medinensis patients at distinct states of infection. Trans R Soc Trop Med Hyg 102: 277–283.
- 25.↑
Laemmli UK, 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685.
- 26.↑
Ko YG, Thompson GA Jr., 1995. Purification of glycosylphosphatidylinositol-anchored proteins by modified triton X-114 partitioning and preparative gel electrophoresis. Anal Biochem 224: 166–172.
- 27.↑
Priest JW, Kwon JP, Moss DM, Roberts JM, Arrowood MJ, Dworkin MS, Juranek DD, Lammie PJ, 1999. Detection by enzyme immunoassay of serum immunoglobulin G antibodies that recognize specific Cryptosporidium parvum antigens. J Clin Microbiol 37: 1385–1392.
- 28.↑
Priest JW, Mehlert A, Arrowood MJ, Riggs MW, Ferguson MA, 2003. Characterization of a low molecular weight glycolipid antigen from Cryptosporidium parvum. J Biol Chem 278: 52212–52222.
- 29.↑
Howe KL et al. 2016. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res 44: D774–D780.
- 30.↑
Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M, 2017. WormBase ParaSite–a comprehensive resource for helminth genomics. Mol Biochem Parasitol 215: 2–10.
- 31.↑
Moss DM, Montgomery JM, Newland SV, Priest JW, Lammie PJ, 2004. Detection of cryptosporidium antibodies in sera and oral fluids using multiplex bead assay. J Parasitol 90: 397–404.
- 32.↑
Priest JW, Moss DM, 2020. Measuring cryptosporidium serologic responses by multiplex bead assay. Methods Mol Biol 2052: 61–85.
- 33.↑
Waterboer T, Sehr P, Pawlita M, 2006. Suppression of non-specific binding in serological Luminex assays. J Immunol Methods 309: 200–204.
- 34.↑
Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H, 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37: 420–423.
- 35.↑
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ, 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402.
- 36.↑
Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schaffer AA, Yu YK, 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272: 5101–5109.
- 37.↑
Zweig MH, Campbell G, 1993. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 39: 561–577.
- 38.
Youden WJ, 1950. Index for rating diagnostic tests. Cancer 3: 32–35.
- 39.↑
Kumar R, Indrayan A, 2011. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr 48: 277–287.
- 40.↑
Zhu XQ et al. 2015. Genetic blueprint of the zoonotic pathogen Toxocara canis. Nat Commun 6: 6145.
- 41.↑
Muller R, 1970. Dracunculus medinensis: diagnosis by indirect fluorescent antibody technique. Exp Parasitol 27: 357–361.
- 42.↑
Prakash D, Parab PB, Sharma RN, 1993. Immunodiagnosis of dracunculiasis by dot-ELISA. Ann Trop Med Parasitol 87: 195–199.
- 43.↑
Garate T, Kliks MM, Cabrera Z, Parkhouse RM, 1990. Specific and cross-reacting antibodies in human responses to Onchocerca volvulus and Dracunculus medinensis infections. Am J Trop Med Hyg 42: 140–147.
- 44.↑
Dissanayake S, Xu M, Piessens WF, 1992. A cloned antigen for serological diagnosis of Wucheraria bancrofti microfilaremia with daytime blood samples. Mol Biochem Parasitol 56: 269–278.
- 45.↑
Chandrashekar R, Curtis KC, Ramzy RM, Liftis F, Li BW, Weil GJ, 1994. Molecular cloning of Brugia malayi antigens for diagnosis of lymphatic filariasis. Mol Biochem Parasitol 64: 261–271.
- 46.↑
Bradley JE, Tuan RS, Shepley KJ, Tree TI, Maizels RM, Helm R, Gregory WF, Unnasch TR, 1993. Onchocerca volvulus: characterization of an immunodominant hypodermal antigen present in adult and larval parasites. Exp Parasitol 77: 414–424.
- 47.↑
Feeser KR, Cama V, Priest JW, Thiele EA, Wiegand RE, Lakwo T, Feleke SM, Cantey PT, 2017. Characterizing reactivity to Onchocerca volvulus antigens in multiplex bead assays. Am J Trop Med Hyg 97: 666–672.
- 48.
Lammie PJ, Weil G, Noordin R, Kaliraj P, Steel C, Goodman D, Lakshmikanthan VB, Ottesen E, 2004. Recombinant antigen-based antibody assays for the diagnosis and surveillance of lymphatic filariasis–a multicenter trial. Filaria J 3: 9.
- 49.↑
Rao KV, Eswaran M, Ravi V, Gnanasekhar B, Narayanan RB, Kaliraj P, Jayaraman K, Marson A, Raghavan N, Scott AL, 2000. The Wuchereria bancrofti orthologue of Brugia malayi SXP1 and the diagnosis of bancroftian filariasis. Mol Biochem Parasitol 107: 71–80.
- 50.↑
Lu J, Holmgren A, 2014. The thioredoxin superfamily in oxidative protein folding. Antioxid Redox Signal 21: 457–470.
- 51.↑
Kunchithapautham K, Padmavathi B, Narayanan RB, Kaliraj P, Scott AL, 2003. Thioredoxin from Brugia malayi: defining a 16-kilodalton class of thioredoxins from nematodes. Infect Immun 71: 4119–4126.
- 52.↑
Ditgen D, Anandarajah EM, Hansmann J, Winter D, Schramm G, Erttmann KD, Liebau E, Brattig NW, 2016. Multifunctional thioredoxin-like protein from the gastrointestinal parasitic Nematodes Strongyloides ratti and Trichuris suis affects mucosal homeostasis. J Parasitol Res 2016: 8421597.
- 53.↑
Krolewiecki AJ et al. 2010. Improved diagnosis of Strongyloides stercoralis using recombinant antigen-based serologies in a community-wide study in northern Argentina. Clin Vaccine Immunol 17: 1624–1630.
- 54.↑
Hamlin KL, Moss DM, Priest JW, Roberts J, Kubofcik J, Gass K, Streit TG, Nutman TB, Eberhard ML, Lammie PJ, 2012. Longitudinal monitoring of the development of antifilarial antibodies and acquisition of Wuchereria bancrofti in a highly endemic area of Haiti. PLoS Negl Trop Dis 6: e1941.